Nomenclature
Short Name:
ZC1
Full Name:
Mitogen-activated protein kinase kinase kinase kinase 4
Alias:
- EC 2.7.11.1
- M4K4
- MAP4K4
- MAPK kinase kinase kinase 4
- MAPK/ERK kinase kinase kinase 4
- MEK kinase kinase 4; MEKKK 4; MEKKK4; Mitogen-activated protein kinase kinase kinase kinase 4; Nck interacting kinase; NIK
- FLH21957
- Hepatocyte progenitor kinase-like/germinal center kinase-like kinase
- HGK
- HPK/GCK-like kinase HGK
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE20
SubFamily:
MSN
Structure
Mol. Mass (Da):
142,101
# Amino Acids:
1239
# mRNA Isoforms:
5
mRNA Isoforms:
150,913 Da (1319 AA; O95819-3); 142,101 Da (1239 AA; O95819); 141,929 Da (1242 AA; O95819-5); 138,418 Da (1211 AA; O95819-2); 133,401 Da (1165 AA; O95819-4)
4D Structure:
Interacts with the SH3 domain of the adapter proteins Nck By similarity. Interacts (via its CNH regulatory domain) with ATL1 (via the N-terminal region).
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
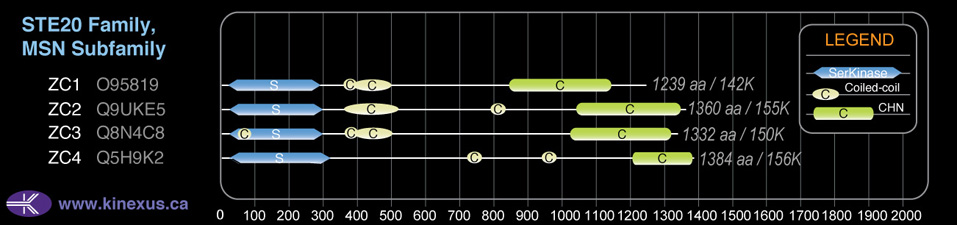
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 25 | 289 | Pkinase |
| 359 | 379 | Coiled-coil |
| 384 | 492 | Coiled-coil |
| 926 | 1213 | CNH |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S5, S175+, S264, S324, S326, S464, S523, S536, S550, S580, S598, S600, S602, S604, S629, S631, S639, S644, S649, S656, S677, S679, S680, S681, S683, S684, S686, S688, S692, S696, S698, S700, S708, S709, S710, S712, S715, S717, S791, S800, S801, S805, S811, S823, S830, S842, S852, S853, S854, S855, S868, S870, S871, S877, S900, S913, S995, S1224.
Threonine phosphorylated:
T35, T59, T124, T181+, T187+, T191-, T319, T351, T576, T627, T628, T809, T828, T834, T840, T846, T857, T873, T874, T876, T907, T915, T1146, T1230.
Tyrosine phosphorylated:
Y36, Y86, Y132, Y193-, Y321, Y323, Y467, Y520, Y560, Y992, Y1143, Y1227.
Ubiquitinated:
K285, K1010, K1198.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1338
50
1117
 2
2
26
21
17
 18
18
239
28
294
 53
53
707
191
794
 61
61
817
55
609
 6
6
76
101
53
 8
8
102
64
269
 62
62
836
68
1580
 30
30
398
20
375
 6
6
82
152
143
 15
15
207
52
332
 53
53
711
215
657
 20
20
274
50
391
 6
6
75
15
56
 10
10
140
47
232
 2
2
29
30
20
 15
15
198
274
1816
 12
12
162
40
227
 11
11
144
142
232
 60
60
806
191
638
 71
71
951
46
1234
 13
13
180
50
226
 17
17
222
30
283
 37
37
493
40
1409
 21
21
278
46
348
 57
57
760
129
1427
 20
20
265
56
387
 10
10
128
40
166
 13
13
180
40
238
 10
10
136
56
180
 57
57
764
24
625
 39
39
526
57
683
 10
10
137
145
244
 65
65
872
130
722
 21
21
282
92
583
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 73.6
73.6
82.9
95 20.8
20.8
29.1
96 -
-
-
93.5 -
-
-
- 63.8
63.8
74.2
93 -
-
-
- 65.4
65.4
75.4
92 25.6
25.6
40.8
94 -
-
-
- -
-
-
- 21
21
29
94 20
20
28.8
93 21.2
21.2
29.8
87 -
-
-
- -
-
-
44 -
-
-
- 49.5
49.5
62.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | NCK1 - P16333 |
| 2 | SLC9A1 - P19634 |
| 3 | GBP3 - Q9H0R5 |
| 4 | RAP2A - P10114 |
| 5 | MAP3K7 - O43318 |
| 6 | ITGB1 - P05556 |
| 7 | DOK1 - Q99704 |
| 8 | CASP8 - Q14790 |
| 9 | BRCA1 - P38398 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
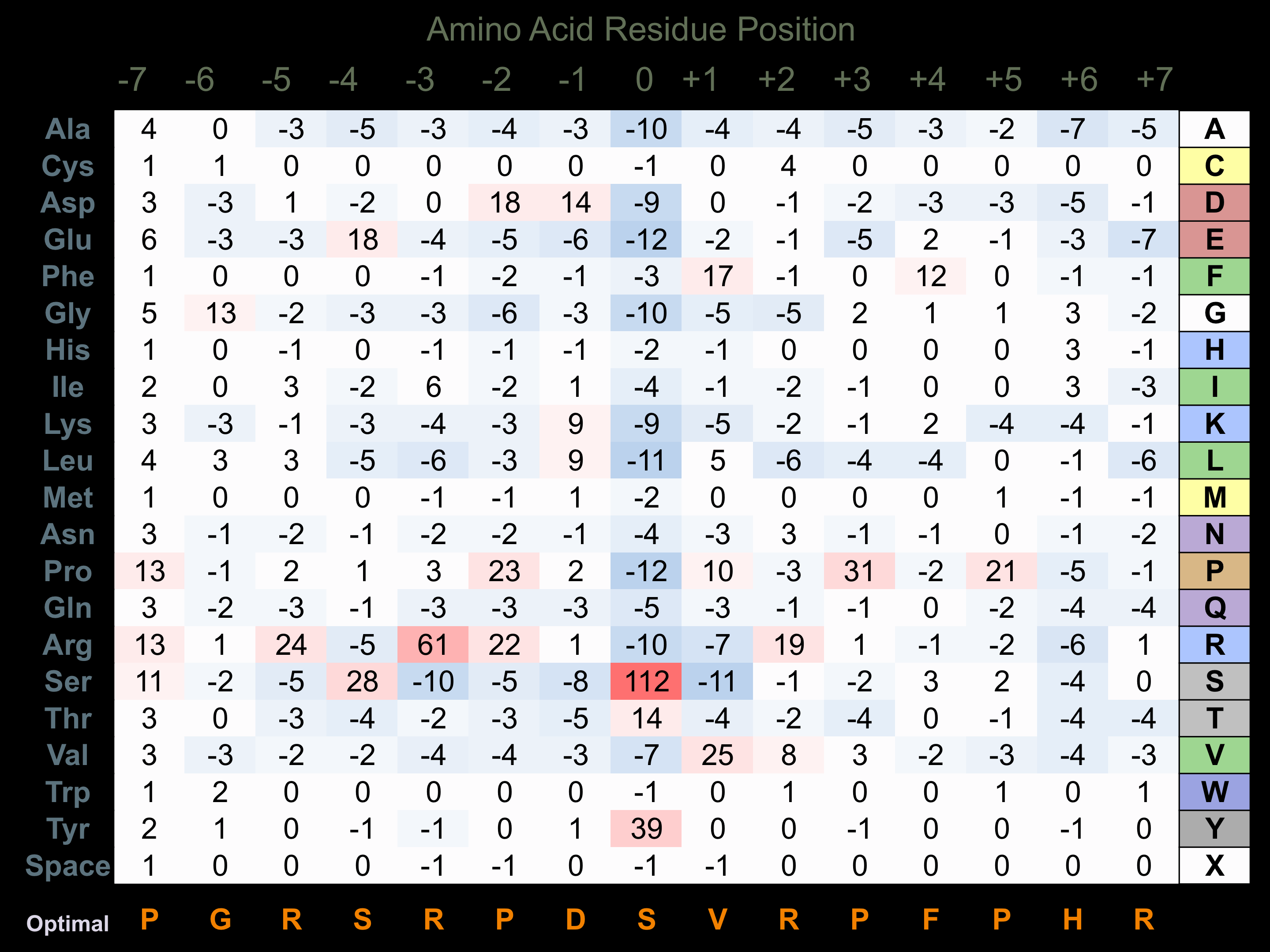
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +96, p<0.003); Clear cell renal cell carcinomas (cRCC) (%CFC= +68, p<0.016); Colorectal adenocarcinomas (early onset) (%CFC= +76, p<0.0002); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +71, p<(0.0003); Skin fibrosarcomas (%CFC= +126, p<0.001); Skin squamous cell carcinomas (%CFC= +74, p<0.008); and Uterine fibroids (%CFC= +110, p<0.0002). The COSMIC website notes an up-regulated expression score for ZC1 in diverse human cancers of 431, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 29 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24998 diverse cancer specimens. This rate is only -12 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.4 % in 1128 large intestine cancers tested; 0.3 % in 807 skin cancers tested; 0.19 % in 590 stomach cancers tested; 0.16 % in 602 endometrium cancers tested; 0.08 % in 1810 lung cancers tested; 0.08 % in 1302 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S259L (6).
Comments:
Only 6 deletions, 2 insertions and no complex mutations are noted on the COSMIC website.

