Nomenclature
Short Name:
ANKRD3
Full Name:
Ankyrin repeat domain protein 3
Alias:
- ANKK2
- PKC-delta-interacting protein kinase
- PKC-regulated kinase PKK
- Receptor-interacting serine-threonine kinase 4
- RIP4
- RIPK4
- Ankyrin repeat domain protein 3
- DIK
- EC 2.7.11.1
- Kinase ANKRD3
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
RIPK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
91,611
# Amino Acids:
832
# mRNA Isoforms:
2
mRNA Isoforms:
91,611 Da (832 AA; P57078); 86,441 Da (784 AA; P57078-2)
4D Structure:
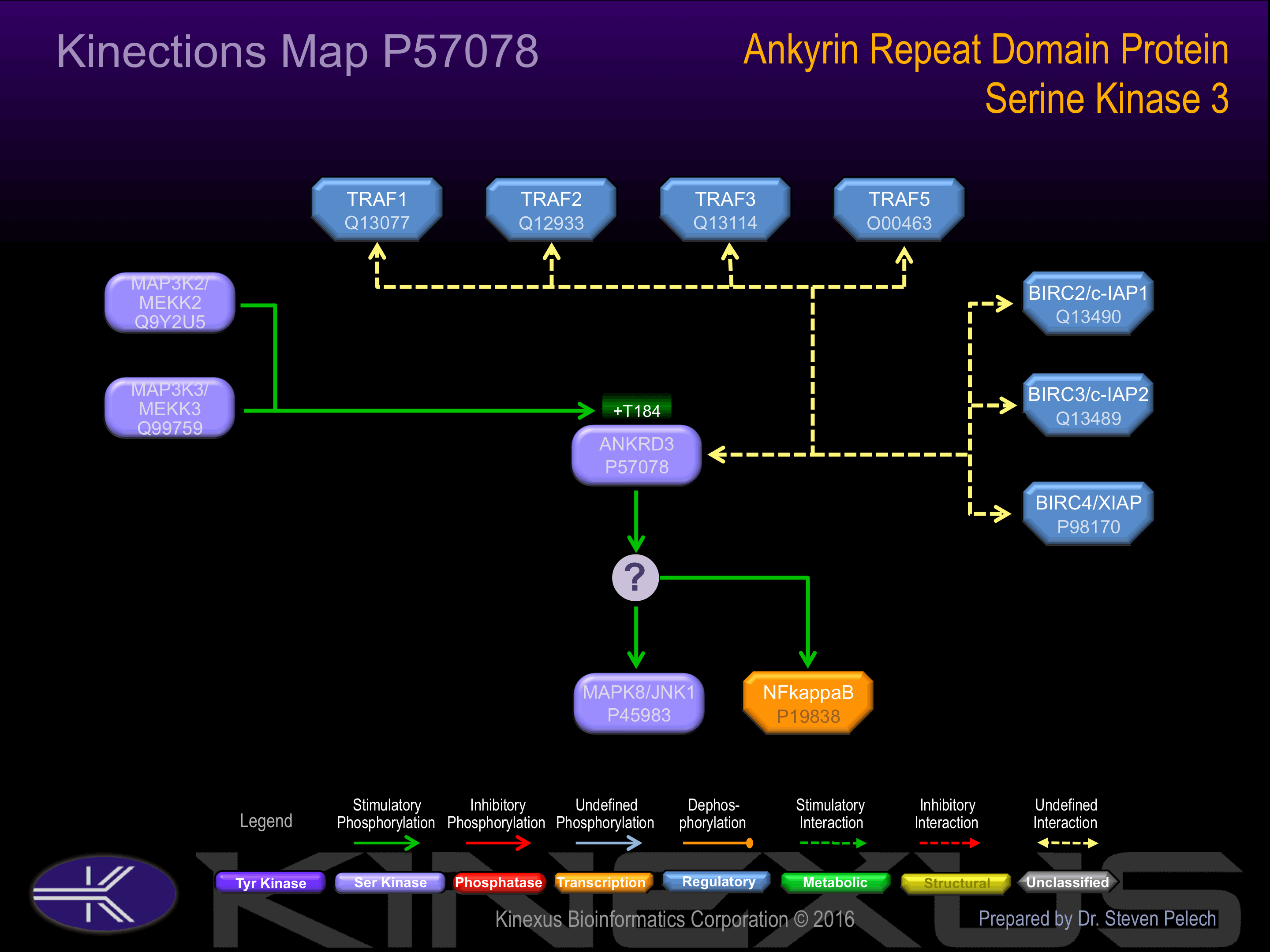
Interacts with PRKCB. Interacts with TRAF1, TRAF2, TRAF3 and TRAF5.
1D Structure:
Subfamily Alignment
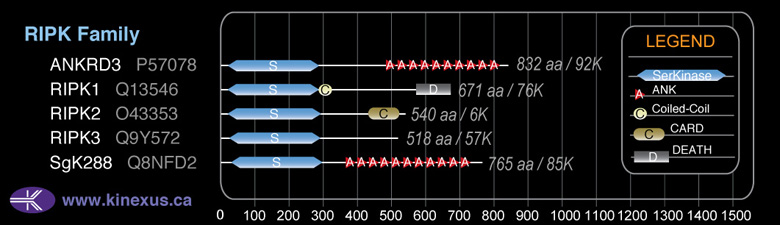
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S415, S420, S427, S438, S444, S520, S544+.
Threonine phosphorylated:
T184+, T521.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 49
49
1487
22
2752
 1
1
31
9
44
 2
2
67
8
85
 26
26
776
74
2741
 27
27
826
22
591
 6
6
189
46
139
 2
2
50
25
37
 77
77
2336
25
5269
 22
22
657
10
566
 5
5
150
64
139
 5
5
166
19
140
 27
27
815
87
743
 1.2
1.2
37
19
59
 0.5
0.5
15
6
13
 4
4
112
16
96
 0.9
0.9
28
12
56
 6
6
176
88
116
 6
6
169
13
206
 1.2
1.2
35
62
42
 28
28
854
84
739
 1.4
1.4
41
15
58
 1.1
1.1
34
17
65
 4
4
106
9
96
 2
2
63
13
90
 6
6
177
15
154
 23
23
689
49
692
 6
6
182
22
115
 8
8
241
13
155
 2
2
69
13
107
 3
3
86
28
53
 19
19
564
18
408
 100
100
3019
26
4987
 0.1
0.1
2
29
2
 24
24
721
52
654
 10
10
307
35
770
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 93.5
93.5
94
99 92.7
92.7
93.5
98.5 -
-
-
90 -
-
-
- 83.3
83.3
88.3
89 -
-
-
- 85
85
89.3
91 23.1
23.1
38.9
91 -
-
-
- 77.5
77.5
85.7
- 72.1
72.1
83.8
77.5 61.7
61.7
73
71 22
22
35.9
63 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 21.4
21.4
35
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PRKCB - P05771 |
Regulation
Activation:
Phosphorylation at Thr-184 increases the phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
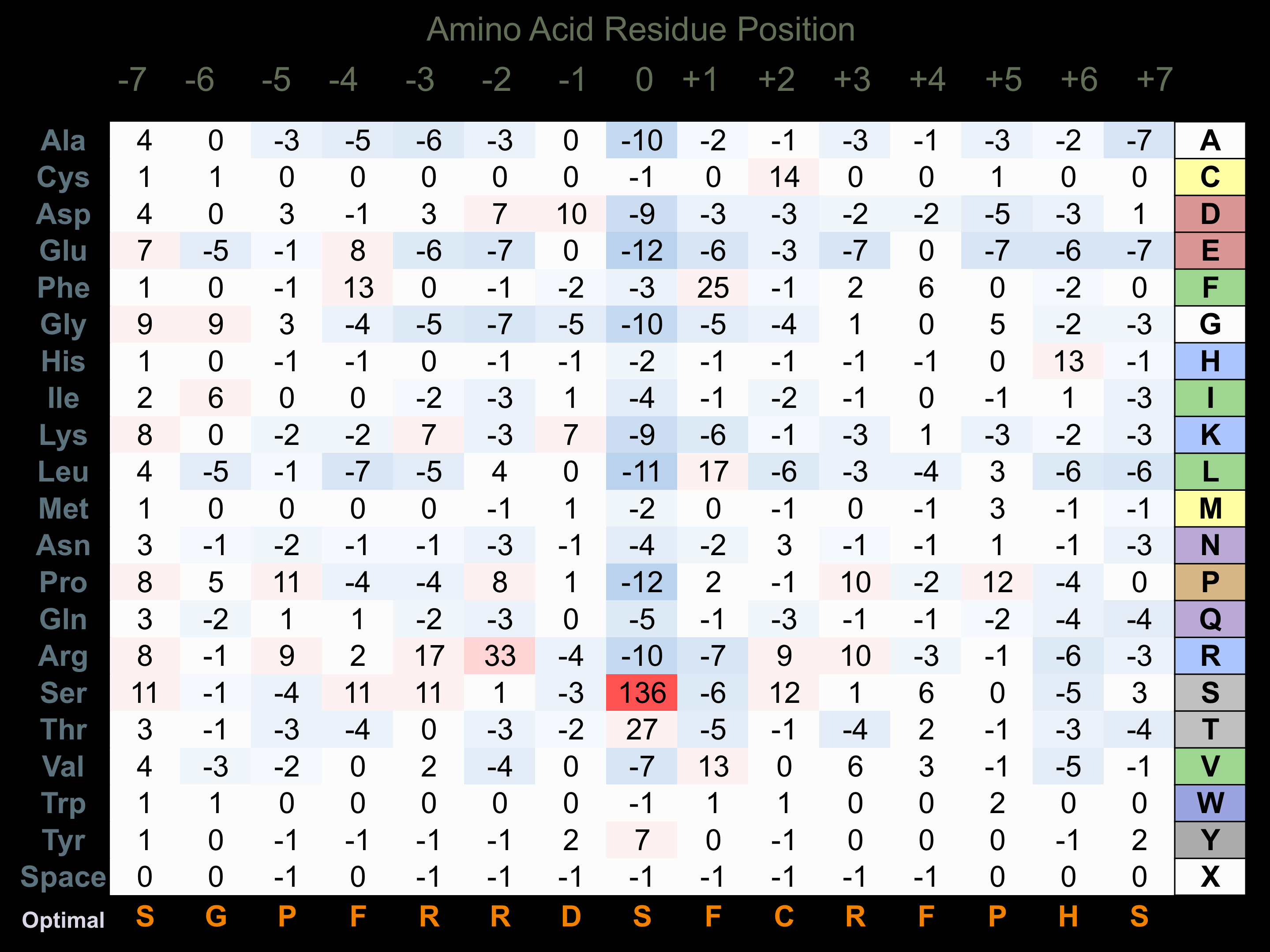
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Lestaurtinib | Kd = 13 nM | 126565 | 22037378 | |
| TG100115 | Kd = 97 nM | 10427712 | 230011 | 22037378 |
| Staurosporine | Kd = 110 nM | 5279 | 22037378 | |
| KW2449 | Kd = 130 nM | 11427553 | 1908397 | 22037378 |
| AST-487 | Kd = 180 nM | 11409972 | 574738 | 22037378 |
| R406 | Kd = 220 nM | 11984591 | 22037378 | |
| PP242 | Kd = 290 nM | 25243800 | 22037378 | |
| Tozasertib | Kd = 300 nM | 5494449 | 572878 | 22037378 |
| Nintedanib | Kd = 440 nM | 9809715 | 502835 | 22037378 |
| Aurora A Inhibitor 1 (DF) | Kd < 600 nM | 21992004 | ||
| Vandetanib | Kd = 620 nM | 3081361 | 24828 | 22037378 |
| Aurora A Inhibitor 23 (DF) | Kd < 800 nM | 21992004 | ||
| Foretinib | Kd = 900 nM | 42642645 | 1230609 | 22037378 |
| BMS-690514 | Kd > 1 µM | 11349170 | 21531814 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Axitinib | Kd = 1.4 µM | 6450551 | 1289926 | 22037378 |
| Crizotinib | Kd = 2.1 µM | 11626560 | 601719 | 22037378 |
| Ruxolitinib | Kd = 2.1 µM | 25126798 | 1789941 | 22037378 |
| Sunitinib | Kd = 2.1 µM | 5329102 | 535 | 19654408 |
| NVP-TAE684 | Kd = 3.2 µM | 16038120 | 509032 | 22037378 |
| Erlotinib | Kd = 4.9 µM | 176870 | 553 | 22037378 |
Disease Linkage
General Disease Association:
Facial, skin, and genital development disorders
Specific Diseases (Non-cancerous):
Popliteal Pterygrium syndrome, Lethal Type (PPS-L); Pterygium, Popliteal Pterygrium syndrome; Severe nonproliferative diabetic retinopathy; Popliteal pterygium syndrome lethal type
Comments:
Loss of function due to I81N, I121N, T184I, and S376X mutations is associated with Popliteal pterygium syndrome, lethal type (PPS-L), which is an autosomal recessive disorder characterized by multiple popliteal pterygia leading to severe arthrogryposis and other symptoms. Early lethality is common.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -59, p<0.015); Bladder carcinomas (%CFC= +110, p<0.008); Breast epithelial carcinomas (%CFC= -59, p<0.093); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -95, p<0.0001); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +97, p<0.035); Skin fibrosarcomas (%CFC= -92); Skin melanomas - malignant (%CFC= -88, p<0.007); and Skin squamous cell carcinomas (%CFC= +68, p<0.003). The COSMIC website notes an up-regulated expression score for ANKRD3 in diverse human cancers of 366, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 27 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 25065 diverse cancer specimens. This rate is only 37 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.56 % in 864 skin cancers tested; 0.39 % in 1270 large intestine cancers tested; 0.24 % in 589 stomach cancers tested; 0.24 % in 1062 upper aerodigestive tract cancers tested; 0.18 % in 548 urinary tract cancers tested; 0.15 % in 238 bone cancers tested; 0.14 % in 603 endometrium cancers tested; 0.13 % in 1459 pancreas cancers tested; 0.09 % in 2082 central nervous system cancers tested; 0.08 % in 710 oesophagus cancers tested; 0.06 % in 1512 liver cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S103F (5); R89C (4).
Comments:
Only 7 deletion, 2 insertions and1 complex mutations are noted on the COSMIC website.