Nomenclature
Short Name:
AurC
Full Name:
Serine-threonine-protein kinase 13
Alias:
- AIE1
- Aurora,Ipl1-related kinase 3
- Aurora-C
- EC 2.7.11.1
- STK13
- STKD
- AIE2
- AIK3
- AURKC
- Aurora,Ipl1,Eg2 protein 1 and 2
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
AUR
SubFamily:
NA
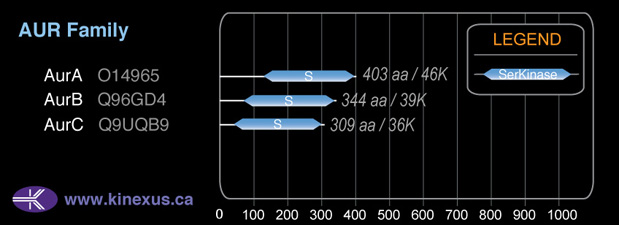
Structure
Mol. Mass (Da):
35,591
# Amino Acids:
309
# mRNA Isoforms:
3
mRNA Isoforms:
35,591 Da (309 AA; Q9UQB9); 33,672 Da (290 AA; Q9UQB9-3); 32,187 Da (275 AA; Q9UQB9-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 43 | 293 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K197.
Serine phosphorylated:
S32, S77, S188+, S271.
Threonine phosphorylated:
T198+, T202-.
Tyrosine phosphorylated:
Y58+, Y205.
Ubiquitinated:
K53, K130.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 47
47
618
16
862
 3
3
45
10
56
 97
97
1293
7
2369
 51
51
675
52
1690
 20
20
260
14
263
 2
2
22
38
30
 24
24
316
19
607
 85
85
1130
25
2100
 14
14
191
10
169
 17
17
230
51
620
 26
26
349
20
711
 48
48
640
90
651
 30
30
398
18
904
 4
4
57
8
51
 20
20
272
17
735
 2
2
20
8
28
 10
10
132
105
571
 28
28
367
14
761
 11
11
142
49
340
 20
20
264
56
286
 32
32
423
16
837
 26
26
350
18
738
 35
35
463
8
673
 55
55
735
14
852
 33
33
438
16
1034
 100
100
1327
32
2584
 16
16
206
21
474
 40
40
536
14
1285
 25
25
334
14
650
 27
27
356
14
112
 25
25
327
18
221
 43
43
574
21
719
 20
20
268
49
510
 62
62
817
26
702
 50
50
666
22
681
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
100
100 63.5
63.5
64.8
95 -
-
-
84 -
-
-
- 70.5
70.5
78.9
88 -
-
-
- 72.5
72.5
82.5
80 67.3
67.3
79.3
79 -
-
-
- 64.7
64.7
78.3
- 52.3
52.3
62.4
- 58.1
58.1
71.2
- 65.3
65.3
78.8
- -
-
-
- 43.2
43.2
62.6
52 -
-
-
- 54.7
54.7
68.9
53 -
-
-
- -
-
-
- -
-
-
- -
-
-
57 56.3
56.3
70.9
57 38.7
38.7
55.3
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
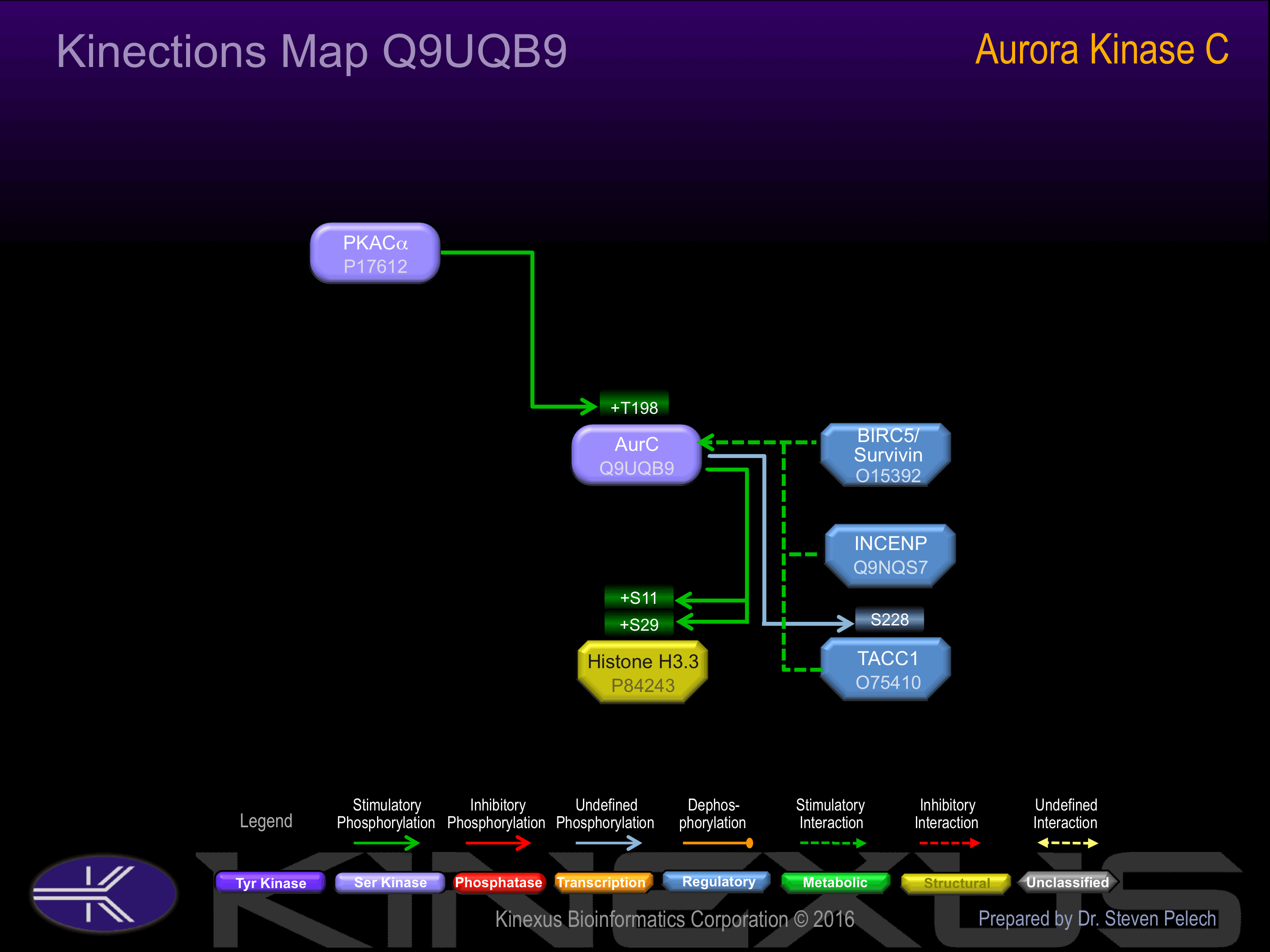
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | INCENP - Q9NQS7 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
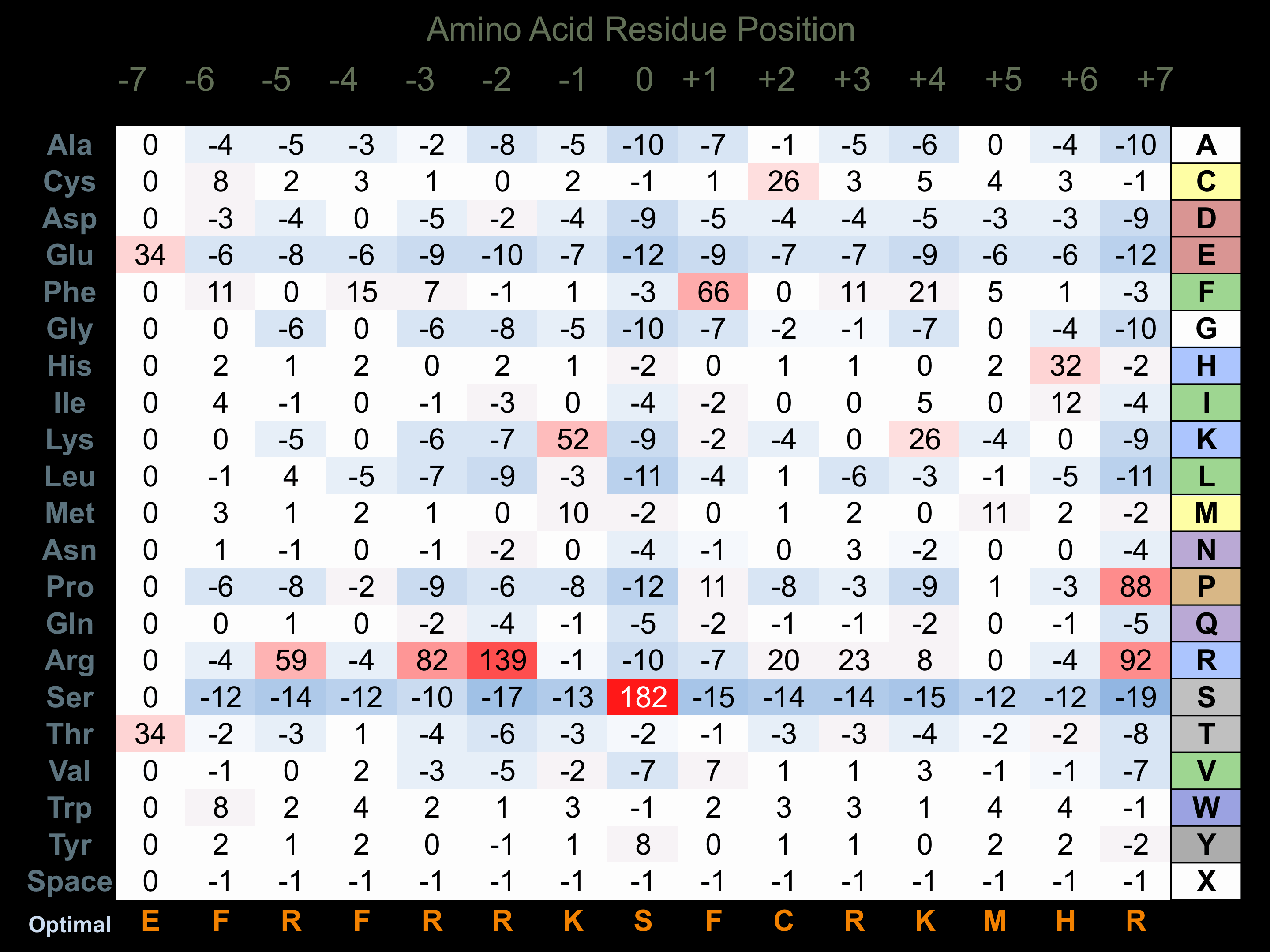
Matrix Type:
Experimentally derived from alignment of 2 known protein substrate phosphosites and 36 peptides phosphorylated by recombinant AurC in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Reproductive disorders
Specific Diseases (Non-cancerous):
Spermatogenic failure 5; Polyploidy
Comments:
AurC has also been shown to play a key role in meiosis, specifically during spermatogenesis production of sperm). Loss-of-function mutations in AURK1 have been linked with spermatogenic failure 5, a male reproductive disorder caused by defective spermatogenesis and usually characterized by the formation of polyploid sperm. Semen from affected males contains multiflagellar spermatozoa with reduced motility, enlarged heads, and abnormalities in the midpiece and acrosome, resulting in infertility. Several mutations in the AurC gene have been observed in male patients with spermatogenic failure 5, including a 1-bp deletion 144delC) leading to premature translation termination at position 71 and a truncated protein without a catalytic domain, a C229Y substitution mutation in the kinase catalytic domain of the protein, and a 2A-to-G transition mutation at the splice site of intron 4 436-2A-G) leading to the omission of exon 5 from the protein. As these mutations result in a loss-of-function for the protein catalytic activity, defects in AurC activity are thought to be the primary contributing factor to the development of spermatogenic failure 5.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -47, p<0.0001); Cervical cancer (%CFC= +131, p<0.0001); Cervical cancer stage 1B (%CFC= +86); Cervical cancer stage 2A (%CFC= +126); Malignant pleural mesotheliomas (MPM) tumours (%CFC= -48, p<0.045); and Prostate cancer - primary (%CFC= +76, p<0.004). The COSMIC website notes an up-regulated expression score for AurC in diverse human cancers of 393, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 25537 diverse cancer specimens. This rate is 1.62-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.87 % in 1270 large intestine cancers tested; 0.6 % in 864 skin cancers tested; 0.6 % in 589 stomach cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,789 cancer specimens
Comments:
Only 1 deletion, no insertions and no complex mutations are noted on the COSMIC website.