Nomenclature
Short Name:
PKACa
Full Name:
cAMP-dependent protein kinase, alpha-catalytic subunit
Alias:
- Alpha-catalytic subunit
- Kinase PKA C-alpha
- PKA C-alpha
- PKA-alpha
- PKACA; Protein kinase, cAMP-dependent, catalytic, alpha
- cAMP-dependent protein kinase
- cAMP-dependent protein kinase, alpha-catalytic subunit
- cAPKa
- EC 2.7.11.11
- KAPCA
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKA
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
40590
# Amino Acids:
351
# mRNA Isoforms:
2
mRNA Isoforms:
40,590 Da (351 AA; P17612); 39,822 Da (343 AA; P17612-2)
4D Structure:
A number of inactive tetrameric holoenzymes are produced by the combination of homo- or heterodimers of the different regulatory subunits associated with two catalytic subunits. cAMP causes the dissociation of the inactive holoenzyme into a dimer of regulatory subunits bound to four cAMP and two free monomeric catalytic subunits.;; C2R2 - a dimer of two catalytic subunits and 2 regulatory subunits. RI (uniprot), RII (uniprot)
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
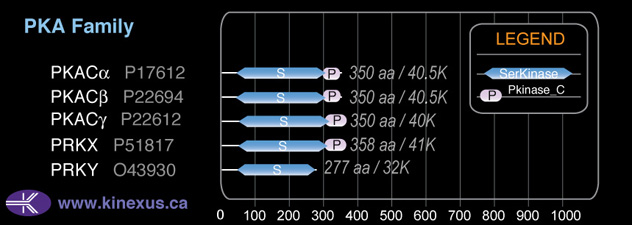
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K267, K280.
Myristoylated:
G2.
Serine phosphorylated:
S11, S15, S54-, S140, S260, S264, S339+.
Threonine phosphorylated:
T49-, T52, T89, T196+, T198+, T202-.
Tyrosine phosphorylated:
Y205, Y331.
Ubiquitinated:
K17, K24, K84, K93, K190, K267, K280, K286, K320.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 81
81
1256
29
1789
 18
18
279
18
287
 21
21
328
11
292
 27
27
412
109
522
 45
45
702
25
607
 84
84
1299
89
3863
 30
30
470
39
639
 57
57
888
49
870
 32
32
494
17
406
 14
14
217
107
232
 15
15
226
36
240
 33
33
512
216
644
 9
9
140
33
173
 7
7
111
13
104
 7
7
106
21
102
 22
22
337
16
405
 7
7
110
290
860
 15
15
240
20
285
 18
18
282
114
389
 30
30
460
109
536
 16
16
254
29
336
 16
16
255
32
306
 16
16
244
29
211
 24
24
369
22
429
 10
10
151
30
182
 100
100
1551
61
2821
 9
9
137
36
177
 12
12
183
21
166
 14
14
222
21
242
 7
7
101
28
73
 22
22
341
24
288
 61
61
952
36
1277
 4
4
62
73
149
 45
45
696
62
672
 24
24
375
35
773
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 81.5
81.5
82
0 51.3
51.3
56.7
- -
-
-
99 -
-
-
98 93.2
93.2
95.4
99 -
-
-
- 98
98
99.4
98 98
98
99.2
98 -
-
-
- -
-
-
- 81.4
81.4
84.7
95 35.3
35.3
54.1
93 34.9
34.9
53.1
89.5 -
-
-
- 82.4
82.4
90.1
82 -
-
-
- 68.6
68.6
78.5
83 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 45.5
45.5
67.1
53 -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by binding of two cAMP molecules to each of the two associated regulatory subunits in the PKA holoenzyme. Binding of cAMP induces dissociation of the two active catalytic subunits. Phosphorylation of Thr-198 increases phosphotransferase activity. Phosphorylation of S338 plays a key role in stabilizing PKA.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Desmin | P17661 | S44 | AGFGSKGSSSSVTSR | |
| Desmin | P17661 | S59 | VYQVSRTSGGAGGLG | |
| Desmin | P17661 | S81 | GTTRTPSSYGAGELL | |
| Desmin | P17661 | T16 | RVSSYRRTFGGAPGF | |
| Desmin | P17661 | T75 | LRASRLGTTRTPSSY | |
| Desmoplakin | P15924 | S2849 | RSGSRRGSFDATGNS | |
| DJ-1 | Q99497 | S155 | KDGLILTSRGPGTSF | |
| DJ-1 | Q99497 | T154 | EKDGLILTSRGPGTS | |
| Doublecortin | O43602 | S128 | TRTLQALSNEKKAKK | |
| DRP1 | O00429 | S637 | VPVARKLSAREQRDC | |
| DUOX1 | Q9NRD9 | S1217 | SHHFRRRSFRGFWLT | |
| DUOX1 | Q9NRD9 | S955 | KDLCRRASYISQDMI | |
| Dynactin-1 | Q14203 | S19 | TPSGSRMSAEASARP | |
| eEF1B | P24534 | S105 | DDIDLFGSDDEEESE | |
| eEF1B | P24534 | S140 | PALVAKSSILLDVKP | |
| eEF1G | P26641 | S298 | DEFKRKYSNEDTLSV | |
| eEF2K | O00418 | S366 | SPRVRTLSGSRPPLL | - |
| eEF2K | O00418 | S500 | RLHLPRASAVALEVQ | |
| eIF2B | P20042 | S218 | RVGTKKTSFVNFTDI | |
| Emerin | P50402 | S49 | ETQRRRLSPPSSSAA | |
| En-2 | P19622 | S276 | LTEQRRQSLAQELSL | |
| eNOS | P29474 | S1177 | TSRIRTQSFSLQERQ | + |
| eNOS | P29474 | S615 | SYKIRFNSISCSDPL | |
| eNOS | P29474 | S633 | WRRKRKESSNTDSAG | |
| eNOS | P29474 | T495 | TGITRKKTFKEVANA | |
| ERa (ESR1) | P03372 | S236 | IDKNRRKSCQACRLR | ? |
| ERa (ESR1) | P03372 | S305 | IKRSKKNSLALSLTA | + |
| ErbB2 (HER2) | P04626 | T686 | QQKIRKYTMRRLLQE | - |
| ERM | P41161 | S367 | PPYQRRGSLQLWQFL | |
| ETV1 (ER81) | P50549 | S191 | HRFRRQLSEPCNSFP | + |
| ETV1 (ER81) | P50549 | S216 | PMYQRQMSEPNIPFP | + |
| ETV1 (ER81) | P50549 | S334 | PTYQRRGSLQLWQFL | |
| Ezrin | P15311 | S66 | LKLDKKVSAQEVRKE | |
| FAK (PTK2) iso5 | Q05397 | S840 | LKPDVRLSRGSIDRE | |
| FETUB | Q9UGM5 | S303 | SKAGPRGSVQYLPDL | |
| FKBP9 | O95302 | S277 | NCERISQSGDFLRYH | |
| FLNA | P21333 | S2152 | TRRRRAPSVANVGSH | |
| FLNA | P21333 | S2523 | VTGPRLVSNHSLHET | |
| FLNA | P21333 | S2615 | HVGSRLYSVSYLLKD | |
| FLNA | P21333 | S343 | NDKNRTFSVWYVPEV | |
| FLNA | P21333 | T2336 | SGDARRLTVSSLQES | |
| FLNC | Q14315 | T2412 | SDDARRLTVTSLQET | |
| FRAT1 | Q92837 | S188 | RLQQRRGSQPETRTG | |
| Fyn | P06241 | S21 | LTEERDGSLNQSSGY | + |
| Ga(13) | Q14344 | T203 | ILLARRPTKGIHEYD | |
| GABBR2 | O75899 | S893 | EHIQRRLSLQLPILH | |
| GAD67 | Q99259 | T91 | RDARFRRTETDFSNL | |
| GANAB | Q14697 | S916 | GSPESRLSFQHDPET | |
| GATA3 | P23771 | S308 | IKPKRRLSAARRAGT | |
| GATA4 | P43694 | S262 | IKPQRRLSASRRVGL | |
| GBF1 | Q92538 | S174 | ICFEMRLSELLRKSA | |
| GCAP2 | Q9UMX6 | S197 | LAQQRRKSAMF____ | |
| GFAP | P14136 | S13 | ITSAARRSYVSSGEM | |
| GFAP | P14136 | S38 | LGPGTRLSLARMPPP | |
| GFAP | P14136 | S8 | MERRRITSAARRSYV | |
| GFAP | P14136 | T7 | _MERRRITSAARRSY | |
| GFAT | Q06210 | S204 | AVGTRRGSPLLIGVR | |
| GFAT | Q06210 | S234 | TARTQIGSKFTRWGS | |
| GHF1 | P28069 | S115 | KQELRRKSKLVEEPI | |
| GHF1 | P28069 | T220 | RKRKRRTTISIAAKD | |
| GIRK1 | P48549 | S385 | NSKERHNSVECLDGL | |
| GLI1 | P08151 | S544 | VSLERRSSSSSSISS | |
| GLI1 | P08151 | S560 | YTVSRRSSLASPFPP | |
| GLI1 | P08151 | S640 | AGVTRRASDPAQAAD | |
| GLI1 | P08151 | T374 | PGCTKRYTDPSSLRK | |
| GLUD1 | P00367 | T206 | EKITRRFTMELAKKG | |
| GluR1 | P42261 | S849 | FCLIPQQSINEAIRT | |
| GluR1 | P42261 | S863 | TSTLPRNSGAGASSG | |
| GluR4 | P48058 | S862 | IRNKARLSITGSVGE | |
| GluR6 | Q13002 | S856 | NAQLEKRSFCSAMVE | |
| GluR6 | Q13002 | S868 | MVEELRMSLKCQRRL | |
| GLUT2 | P11168 | S491 | VPETKGKSFEEIAAE | |
| GLUT2 | P11168 | S503 | AAEFQKKSGSAHRPK | |
| GLUT2 | P11168 | S505 | EFQKKSGSAHRPKAA | |
| GLUT4 | P14672 | S488 | AAFHRTPSLLEQEVK | |
| GMFb | P60983 | S82 | QHDDGRVSYPLCFIF | |
| GMFb | P60983 | T26 | KFRFRKETNNAAIIM | |
| GNMT | Q14749 | S9 | DSVYRTRSLGVAAEG | |
| GPLD1 | P80108 | T309 | NDFHRNLTTSLTESV | - |
| GRK7 | Q8WTQ7 | S23 | YLQARKPSDCDSKEL | - |
| GRK7 | Q8WTQ7 | S36 | ELQRRRRSLALPGLQ | - |
| GSK3a | P49840 | S21 | SGRARTSSFAEPGGG | - |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| GSTA4 | O15217 | S189 | QEYTVKLSNIPTIKR | |
| GSTP1 | P09211 | S184 | SAYVGRLSARPKLKA | |
| GYS1 | P13807 | S8 | MPLNRTLSMSSLPGL | - |
| GYS2 | P54840 | S8 | MLRGRSLSVTSLGGL | - |
| H-Ras-1 | P01112 | S177 | KLNPPDESGPGCMSC | |
| H11 (HSPB8) | Q9UJY1 | S57 | DWALPRLSSAWPGTL | ? |
| H1D | P16403 | S36 | GGTPRKASGPPVSEL | |
| H1F1 | Q02539 | T132 | VATKTKATGASKKLK | |
| H1R | P35367 | S398 | WKRLRSHSRQYVSGL | |
| H2B | P33778 | S33 | DGKKRKRSRKESYSI | |
| H2B | P33778 | S37 | RKRSRKESYSIYVYK | |
| H3.1 | P68431 | S11 | TKQTARKSTGGKAPR | + |
| H3.2 | P84228 | S11 | TKQTARKSTGGKAPR | |
| H3.2 | P84228 | S29 | ATKAARKSAPATGGV | |
| HDAC8 | Q9BY41 | S39 | AKIPKRASMVHSLIE | |
| HePTP | P35236 | S44 | RLQERRGSNVALMLD | - |
| HMG-CoA reductase | P04035 | S872 | SHMIHNRSKINLQDL | |
| HMGN1 (HMG14) | P05114 | S7 | _PKRKVSSAEGAAKE | |
| hnRNP A1 | P09651 | S199 | SQRGRSGSGNFGGGR | - |
| hnRNP D | Q14103 | S87 | SNSSPRHSEAATAQR | |
| hnRNP I | P26599 | S16 | AVGTKRGSDELFSTC | |
| HPK1 (MAP4K1) | Q92918 | S171 | ATLARRLSFIGTPYW | + |
| HSL | Q05469 | S565 | ASATVRVSRLLSLPP | |
| HSL | Q05469 | S853 | IAEPMRRSVSEAALA | |
| HSL | Q05469 | S950 | EGFHPRRSSQGATQM | |
| HSL | Q05469 | S951 | GFHPRRSSQGATQMP | |
| HSP20 | O14558 | S16 | PSWLRRASAPLPGLS | ? |
| HSP27 | P04792 | S15 | FSLLRGPSWDPFRDW | ? |
| HSP27 | P04792 | S78 | PAYSRALSRQLSSGV | + |
| HSP27 | P04792 | S82 | RALSRQLSSGVSEIR | ? |
| HSP27 | P04792 | T143 | RCFTRKYTLPPGVDP | |
| HSP90AA1 | P07900 | S211 | KEIVKKHSQFIGYPI | |
| HSP90AA1 | P07900 | S263 | PEIEDVGSDEEEEKK | |
| HSP90B | P08238 | S452 | STNRRRLSELLRYHT | |
| ICLN | P54105 | S45 | YIAESRLSWLDGSGL | |
| IP3R1 (IP3 receptor) | Q14643 | S1598 | RNAARRDSVLAASRD | |
| IP3R1 (IP3 receptor) | Q14643 | S1764 | RPSGRRESLTSFGNG | |
| IP3R3 | Q14573 | S1832 | TTKGRVASFSIPGSS | |
| IP3R3 | Q14573 | S916 | GGKNVRRSIQGVGHM | |
| IP3R3 | Q14573 | S934 | MVLSRKQSVFSAPSL | |
| ITCH | Q96J02 | S450 | GWEKRTDSNGRVYFV | |
| ITCH | Q96J02 | T464 | VNHNTRITQWEDPRS | |
| ITGA4 | P13612 | S1027 | QEENRRDSWSYINSK | |
| ITGB4 | P16144 | S1364 | PSGSQRPSVSDDTGC | |
| ITIH4 | Q14624 | S225 | TLSQQQKSPEQQETV | |
| KCNJ13 | O60928 | S287 | EICQRRTSYLPSEIM | |
| KCNJ8 | Q15842 | S385 | NSLRKRNSMRRNNSM | |
| KCNJ8 | Q15842 | T234 | IQVVKKTTTPEGEVV | |
| KCNK18 | Q7Z418 | S252 | QAMERSNSCPELVLG | |
| KCNN2 | Q9H2S1 | S135 | FEKRKRLSDYALIFG | |
| KCNN2 | Q9H2S1 | S464 | QAIHQLRSVKMEQRK | |
| KCNN2 | Q9H2S1 | S567 | SSSRRRRSSSTAPPT | |
| KCNN2 | Q9H2S1 | S568 | SSRRRRSSSTAPPTS | |
| KCNN2 | Q9H2S1 | S569 | SRRRRSSSTAPPTSS | |
| KCNN4 | O15554 | S314 | YTKEMKESAARVLQE | |
| KCNN4 | O15554 | S334 | KHTRRKESHAARRHQ | |
| KCNN4 | O15554 | T329 | AWMFYKHTRRKESHA | |
| KDELR1 | P24390 | S209 | VLKGKKLSLPA____ | |
| Kir5.1 | Q9NPI9 | S416 | LNRISMESQM_____ | |
| Kir6.2 | Q14654 | T224 | MQVVRKTTSPEGEVV | |
| Kv1.2 | P16389 | S449 | PDLKKSRSASTISKS | |
| Kv1.4 | P22459 | S227 | FFDRNRPSFDAILYY | |
| Kv1.4 | P22459 | T601 | LKKFRSSTSSSLGDK | |
| Kv4.2 | Q9NZV8 | T38 | PRQERKRTQDALIVL | |
| L-plastin | P13796 | S5 | ___MARGSVSDEEMM | |
| Lamin A,C | P02545 | S5 | ___METPSQRRATRS | |
| Lasp-1 | Q14847 | S146 | MEPERRDSQDGSSYR | |
| Lasp-1 | Q14847 | S99 | KNKGKGFSVVADTPE | |
| Lck | P06239 | S42 | TLLIRNGSEVRDPLV | |
| LKB1 (STK11) | Q15831 | S428 | SSKIRRLSACKQQ__ | + |
| LPH (Lactase-phlorizin hydrolase) | P09848 | S1917 | KQGKTQRSQQELSPV | |
| LPP | Q93052 | S116 | TLEERRSSLDAEIDS | |
| LRP1 | Q07954 | S4520 | GGHGSRHSLASTDEK | |
| LRPAP1 | P30533 | S242 | LDRLRRVSHQGYSTE | |
| MAP2 iso3 | P11137 | T1577 | SSSARRTTRSEPIRR | |
| MAPKAPK5 | Q8IW41 | S115 | GELFHRISQHRHFTE | ? |
| MARCKS | P29966 | S159 | KKKKKRFSFKKSFKL | |
| MARK3 | P27448 | T507 | GGMTRRNTYVCSERT | |
| MAZ | P56270 | S186 | ALEKKTKSKGPYICA | |
| MAZ | P56270 | T385 | KDRLRAHTVRHEEKV | |
| MC4R | P32245 | T312 | RSQELRKTFKEIICC | |
| MCAM | P43121 | S85 | VHQGKGQSEPGEYEH | |
| MCAM | P43121 | S93 | GEYEQRLSLQDRGAT | |
| MEF2C | Q06413 | S121 | EDKYRRASEELDGLF | |
| MEF2C | Q06413 | S190 | QPALQRNSVSPGLPQ | |
| MEF2C | Q06413 | T20 | DERNRQVTFTKRKFG | + |
| Merlin | P35240 | S10 | GAIASRMSFSSLKRK | |
| Merlin | P35240 | S518 | DTDMKRLSMEIEKEK | |
| mGluR2 | Q14416 | S675 | AQRPRFISPASQVAI | |
| mGluR2 | Q14416 | S843 | GSAAARASSSLGQGS | |
| mGluR3 | Q14832 | S843 | RLHLNRFSVSGTGTT | |
| mGluR7 | Q14831 | S862 | NVQKRKRSFKAVVTA | |
| Moesin | P26038 | T558 | LGRDKYKTLRQIRQG | |
| Mos | P00540 | S25 | SVDARPCSSPSELPA | - |
| MRLC1 (MYL9) | P24844 | S20 | KRPQSATSNVFAMFD | |
| MRLC1 (MYL9) | P24844 | T19 | KKRPQSATSNVFAMF | |
| MST3 (STK24) | Q9Y6E0 | T18 | ALNKRRATLPHPGGS | + |
| Mucolipin 1 | Q9GZU1 | S557 | SGKFRRGSGSACSLL | |
| Mucolipin 1 | Q9GZU1 | S559 | KFRRGSGSACSLLCC | |
| Myb | P10242 | S116 | KYGPKRWSVIAKHLK | |
| MYBPC3 | Q14896 | T274 | LLSAFRRTSLAGGGR | |
| Myf-6 | P23409 | S220 | SSSLRCLSSIVDSIS | |
| Myf-6 | P23409 | S221 | SSLRCLSSIVDSISS | |
| Myf-6 | P23409 | S89 | CKTCKRKSAPTDRRK | |
| MYH11 | P35749 | T1763 | SDRVRKATQQAEQLS | |
| MYLK1 (smMLCK) | Q15746 | S1388 | LATCRSTSFNVQDLL | |
| MYLK1 (smMLCK) | Q15746 | S1760 | RAIGRLSSMAMISGL | |
| MYLK1 (smMLCK) | Q15746 | S1773 | GLSGRKSSTGSPTSP | |
| MYLK1 (smMLCK) | Q15746 | S1776 | GRKSSTGSPTSPINA | |
| MYLK1 (smMLCK) | Q15746 | S1779 | SSTGSPTSPLNAEKL | |
| MYLK1 (smMLCK) | Q15746 | S815 | SLMLQNSSARALPRG | - |
| Myomesin 1 | P52179 | S482 | ARLKSPLSTLDWTVI | |
| Myomesin 2 | P54296 | S76 | RVCAKRVSTQEDEEQ | |
| MYPT1 | O14974 | S695 | QARQSRRSTQGVTLT | |
| MYPT1 | O14974 | S852 | RPREKRRSTGVSFWT | |
| nAChRA4 | P43681 | S362 | LLLMKRPSVVKDNCR | |
| nAChRA4 | P43681 | S467 | LAKARSLSVQHMSSP | |
| nAChRA4 | P43681 | S486 | EGGVRCRSRSIQYCV | |
| nAChRA7 | P36544 | S365 | QHKQRRCSLASVEMS | |
| NCOA3 (SRC-3) | Q9Y6Q9 | S857 | PPYNRAVSLDSPVSV | + |
| NDRG1 | Q92597 | S330 | LMRSRTASGSSVTSL | |
| NDRG1 | Q92597 | T366 | GTRSRSHTSEGAHLD | |
| NDUFS3 | O75489 | S59 | DVAHKQLSAFGEYVA | |
| NEDD4L | Q96PU5 | S342 | SSRLRSCSVTDAVAE | - |
| NEDD4L | Q96PU5 | S448 | IRRPRSLSSPTVTLS | ? |
| NEDD4L | Q96PU5 | T367 | AGRARSSTVTGGEEP | |
| Neurabin 1 | Q9ULJ8 | S460 | ANRKIKFSSAPIKVF | |
| NF-E2 | Q16621 | S170 | TEAGRRRSEYVEMYP | |
| NFAT2 | O95644 | S245 | PSTSPRASVTEESWL | |
| NFAT2 | O95644 | S269 | PCNKRKYSLNGRQPP | |
| NFAT2 | O95644 | S294 | PHGSPRVSVTDDSWL | |
| NFE2L1 | Q14494 | S599 | PSALKKGSKEKQADF | |
| NFkB-p105 | P19838 | S337 | FVQLRRKSDLETSEP | |
| NFkB-p65 | Q04206 | S276 | SMQLRRPSDRELSEP | |
| NFL (Neurofilament L) | P07196 | S13 | YEPYYSTSYKRRYVE | |
| NFL (Neurofilament L) | P07196 | S3 | MSSFSYEPYY_____ | |
| NFL (Neurofilament L) | P07196 | S42 | TARSAYSSYSAPVSS | |
| NFL (Neurofilament L) | P07196 | S435 | QTSSYLMSTRSFPSY | |
| NFL (Neurofilament L) | P07196 | S50 | YSAPVSSSLSVRRSY | |
| NFM (Neurofilament M) | P07197 | S1 | _______SYTLDSLG | |
| NFM (Neurofilament M) | P07197 | S22 | RVTETRSSFSRVSGS | |
| NFM (Neurofilament M) | P07197 | S45 | SWSRGSPSTVSSSYK | |
| NFM (Neurofilament M) | P07197 | S614 | EKPEKAKSPVPKSPV | |
| NFM (Neurofilament M) | P07197 | S619 | AKSPVPKSPVEEKGK | |
| NFM (Neurofilament M) | P07197 | S735 | PEKKKAESPVKEEAV | |
| NHE3 | P48764 | S555 | AEGERRGSLAFIRSP | |
| NHE3 | P48764 | S607 | SLEQRRRSIRDAEDM | |
| NHERF | O14745 | S76 | ETHQQVVSRIRAALN | |
| NIPP-1 | Q12972 | S178 | TAHNKRISTLTIEEG | |
| NIPP-1 | Q12972 | S199 | PKRKRKNSRVTFSED | |
| NMDAR1 | Q05586 | S897 | SFKRRRSSKDTSTGG | |
| NMDAR2C | Q14957 | S1227 | PCTWRRISSLESEV | |
| NOLC1 | Q14978 | S622 | KKGEKRASSPFRRVR | |
| NOLC1 | Q14978 | S623 | KGEKRASSPFRRVRE | |
| NR5A2 | O00482 | S510 | LPEIRAISMQAEEYL | + |
| Nur77 | P22736 | S341 | KEVVRTDSLKGRRGR | |
| Nur77 | P22736 | S351 | GRRGRLPSKPKQPPD | - |
| p47phox | P14598 | S320 | QRSRKRLSQDAYRRN | ? |
| p47phox | P14598 | S328 | QDAYRRNSVRFLQQR | + |
| p47phox | P14598 | S359 | EERQTQRSKPQPAVP | + |
| p47phox | P14598 | S370 | PAVPPRPSADLILNR | + |
| p53 | P04637 | S378 | SKKGQSTSRHKKLMF | + |
| PAH | P00439 | S16 | PGLGRKLSDFGQETS | |
| PAK1 | Q13153 | T423 | PEQSKRSTMVGTPYW | + |
| PAWR | Q96IZ0 | T163 | LREKRRSTGVVNIPA | |
| PCTAIRE1 (CDK16, PCTK1) | Q00536 | S110 | NHPPRKISTEDINKR | |
| PCTAIRE1 (CDK16, PCTK1) | Q00536 | S119 | EDINKRLSLPADIRL | - |
| PCTAIRE1 (CDK16, PCTK1) | Q00536 | S12 | KKIKRQLSMTLRGGR | |
| PCTAIRE1 (CDK16, PCTK1) | Q00536 | S153 | SRRLRRVSLSEIGFG | - |
| PCTAIRE3 (CDK18, PCTK3) | Q07002 | S12 | KNFKRRFSLSVPRTE | + |
| PCTAIRE3 (CDK18, PCTK3) | Q07002 | S130 | SRMSRRASLSDIGFG | + |
| PCTAIRE3 (CDK18, PCTK3) | Q07002 | S470 | RGKNRRQSIF_____ | ? |
| PCTAIRE3 (CDK18, PCTK3) | Q07002 | S87 | RQNQRRFSMEDVSKR | + |
| PDE10A iso2 | Q9Y233-2 | T16 | ASCFRRLTECFLSPS | ? |
| PDE1A | P54750 | S119 | EEKPKFRSIVHAVQA | |
| PDE1A | P54750 | T137 | VERMYRKTYHMVGLA | |
| PDE3A | Q14432 | S312 | SKSHRRTSLPCIPRE | |
| PDE3B | Q13370 | S296 | IRPRRRSSCVSLGET | |
| PDE3B | Q13370 | S318 | CKIFRRPSLPCISRE | |
| PDE3B | Q13370 | S73 | PQQPRRCSPFCRARL | |
| PDE4A | P27815 | S145 | ATSQRRESFLYRSDS | + |
| PDE4B | Q07343 | S133 | GHSQRRESFLYRSDS | + |
| PDE4C | Q08493 | S119 | PHSQRRESFLYRSDS | + |
| PDE4D iso2 | Q08499-2 | S12 | NNFFRRHSWICFDVD | |
| PDE4D iso6 | Q08499-6 | S126 | VHSQRRESFLYRSDS | |
| PDE5A | O76074 | S102 | GTPTRKISASEFDRP | + |
| PDE6G | P18545 | T35 | PKFKQRQTRQFKSKP | |
| PDIA6 | Q15084 | S375 | KFALLKGSFSEQGIN | |
| PDIA6 | Q15084 | S88 | VDADKHHSLGGQYGV | |
| PDZK1 | Q5T2W1 | S505 | MAKERAHSTASHSSS | |
| PEDF | P36955 | S227 | KFDSRKTSLEDFYLD | |
| PEDF | P36955 | T226 | TKFDSRKTSLEDFYL | |
| perilipin | O60240 | S223 | PSLLRRVSTLANTLS | |
| perilipin | O60240 | S277 | QAVSRRRSEVRVPWL | |
| perilipin | O60240 | S497 | EKPKRRVSDSFFRPS | |
| perilipin | O60240 | S522 | YSQLRKKS_______ | |
| Perilipin | O60240 | S81 | EPVVRRLSTQFTAAN | |
| PFKFB1 | P16118 | S33 | RLQRRRGSSIPQFTN | |
| PFKFB2 | O60825 | S466 | PVRMRRNSFTPLSSS | |
| PFKFB2 | O60825 | S483 | IRRPRNYSVGSRPLK | |
| PFKFB3 | Q16875 | S461 | NPLMRRNSVTPLASP | |
| PFKL | P17858 | T773 | LEHVTRRTLSMDKGF | |
| PFKM | P08237 | S774 | EHITRKRSGEAAV__ | |
| PFKP | Q01813 | S12 | DSRAPKGSLRKFLEH | |
| PGM5 | Q15124 | T31 | GGGLRRPTGLFEGQR | |
| PHKg1 | Q16816 | S81 | VDILRKVSGHPNIIQ | |
| PHKg1 | Q16816 | Y337 | EIVIRDPYALRPLRR | |
| Phosducin | P20941 | S73 | ERVSRKMSIQEYELI | |
| Phospholamban | P26678 | S16 | RSAIRRASTIEMPQQ | |
| Phospholamban | P26678 | T17 | SAIRRASTIEMPQQA | |
| PIK3R1 | P27986 | S83 | YIGRKKISPPTPKPR | + |
| Pin1 | Q13526 | S16 | PGWEKRMSRSSGRVY | - |
| PIP5K1B | O14986 | S214 | STYKRRASRKEREKS | - |
| PKACa (PRKACA) | P17612 | S11 | AAAAKKGSEQESVKE | |
| PKACa (PRKACA) | P17612 | S140 | LRRIGRFSEPHARFY | |
| PKACa (PRKACA) | P17612 | S339 | EEEEIRVSINEKCGK | + |
| PKACa (PRKACA) | P17612 | T198 | RVKGRTWTLCGTPEY | + |
| PKAR2A | P13861 | S98 | SRFNRRVSVCAETYN | |
| PKAR2A | P13861 | S99 | SRFNRRVSVCAETYN | |
| PKAR2B | P31323 | S114 | NRFTRRASVCAEAYN | |
| PLCB3 | Q01970 | S1105 | LDRKRHNSISEAKMR | - |
| PLCG1 | P19174 | S1248 | HGRAREGSFESRYQQ | |
| PLCL1 | Q15111 | S95 | GGRKKTVSFSSMPSE | |
| PLCL1 | Q15111 | T246 | PRFMWLKTVFEAADV | |
| PLCL1 | Q15111 | T93 | KCGGRKKTVSFSSMP | |
| PLM | O00168 | S83 | EEGTFRSSIRRLSTR | |
| PLM | O00168 | S88 | RSSIRRLSTRRR___ | |
| polycystin 1 | P98161 | S4169 | PSRSSRGSKVSPDVP | |
| Polycystin 1 | P98161 | S4252 | SLQGRRSSRAPAGSS | |
| POU2F1 | P14859 | S385 | RRRKKRTSIETNIRV | - |
| PPAR-binding protein | Q15648 | S18 | EKLSKMSSLLERLHA | |
| PPAR-binding protein | Q15648 | T796 | EASKLPSTSDDCPAI | |
| PPP1R14A (CPI 17) | Q96A00 | S130 | GLRQPSPSHDGSLSP | |
| PPP1R14A (CPI 17) | Q96A00 | T38 | QKRHARVTVKYDRRE | - |
| PPP1R16B | Q96T49 | S337 | SSLSRRTSSAGSRGK | |
| PPP1R1A | Q13522 | S6 | __MEQDNSPRKIQFT | |
| PPP1R1A | Q13522 | S67 | LKSTLAMSPRQRKKM | |
| PPP1R1A | Q13522 | T35 | QIRRRRPTPATLVLT | |
| PPP1R1A | Q13522 | T75 | PRQRKKMTRITPTMK | + |
| PPP1R1C | Q8WVI7 | T34 | QIRKRRPTPASLVIL | |
| PPP1R3A | Q16821 | S46 | PQPSRRGSDSSEDIY | |
| PPP1R3A | Q16821 | S65 | SSGTRRVSFADSFGF | |
| PPP2R5D | Q14738 | S573 | KVLLRRKSELPQDVY | |
| PPP2R5D | Q14738 | S60 | PSSNKRPSNSTPPPT | |
| PPP2R5D | Q14738 | S75 | QLSKIKYSGGPQIVK | |
| PPP2R5D | Q14738 | S88 | VKKERRQSSSRFNLS | |
| prolactin | P01236 | S207 | LHCLRRDSHKIDNYL | |
| PSEN1 | P49768 | S310 | PEAQRRVSKNSKYNA | |
| PSMC5 | P62195 | S120 | RVALRNDSYTLHKIL | |
| PTHR | Q03431 | S473 | VQAEIKKSWSRWTLA | |
| PTHR | Q03431 | S475 | AEIKKSWSRWTLALD | |
| PTHR | Q03431 | S491 | KRKARSGSSSYSYGP | |
| PTPN12 (PTP-PEST) | Q05209 | S39 | FMRLRRLSTKYRTEK | - |
| PTPN12 (PTP-PEST) | Q05209 | S435 | KKLERNLSFEIKKVP | |
| PTRF | Q6NZI2 | S300 | SRDKLRKSFTPDHVV | |
| PTRF | Q6NZI2 | S365 | AGDLRRGSSPDVHAL | |
| PTRF | Q6NZI2 | S366 | GDLRRGSSPDVHALL | |
| PTRF | Q6NZI2 | T302 | DKLRKSFTPDHVVYA | |
| Rabphilin 3A | Q9Y2J0 | S237 | GPPVRRASEARMSSS | |
| Rabphilin 3A | Q9Y2J0 | S272 | AGLRRANSVQASRPA | |
| Raf1 | P04049 | S233 | VSSQHRYSTPHAFTF | + |
| Raf1 | P04049 | S259 | SQRQRSTSTPNVHMV | - |
| Raf1 | P04049 | S43 | FGYQRRASDDGKLTD | - |
| Raf1 | P04049 | S621 | PKINRSASEPSLHRA | + |
| Rap1a | P62834 | S180 | KKKPKKKSCLLL___ | |
| Rap1b | P61224 | S179 | PGKARKKSSCQLL__ | |
| RARA | P10276 | S369 | YVRKRRPSRPHMFPK | - |
| RasGRF1 | Q13972 | S759 | SRRRKLISLNIPIIT | |
| RasGRF1 | Q13972 | S835 | PSALSKQSSEVSMRE | |
| RasGRF1 | Q13972 | S929 | KEKYRRMSLASAGFP | |
| Rb | P06400 | S780 | STRPPTLSPIPHIPR | - |
| RCN3 | Q96D15 | S117 | QQRHIRDSVSAAWDT | |
| Ret | P07949 | S696 | SSGARRPSLDSMENQ | + |
| RFC1 | P35251 | S366 | SSPAKKESVSPEDSE | |
| RFC1 | P35251 | T245 | TKKARKDTEAGETFS | |
| RGS10 | O43665 | S176 | QTAAKRASRIYNT__ | |
| RGS4 | P49798 | S52 | VVICQRVSQEEVKKW | |
| RhoA | P61586 | S188 | ARRGKKKSGCLVL__ | |
| RHOK | Q15835 | S21 | AFIAARGSFDGSSSQ | - |
| RIL | P50479 | S120 | PTTSRRPSGTGTGPE | |
| RIMS1 | Q86UR5 | S391 | ARHERRHSDVALPRT | |
| ROMK | P48048 | S219 | RVANLRKSLLIGSHI | |
| ROMK | P48048 | S313 | ATCQVRTSYVPEEVL | |
| ROMK | P48048 | S44 | RQRARLVSKDGRCNI | |
| RPN1 | P04843 | S68 | GSTSRATSFLLALEP | |
| RPN1 | P04843 | T67 | GGSTSRATSFLLALE | |
| RPS19 | P39019 | S59 | WFYTRAASTARHLYL | |
| RPS19 | P39019 | S90 | QRNGVMPSHFSRGSK | |
| RRAD | P55042 | S273 | AGTRRRESLGKKAKR | |
| RXRa | P19793 | S27 | TSPTGRGSMAAPSLH | - |
| RyR1 | P21817 | S2843 | KKKTRKISQSAQTYD | |
| RyR2 | Q92736 | S2029 | TIRGRLLSLVEKVTY | |
| RyR2 | Q92736 | S2806 | YNRTRRISQTSQVSV | |
| S6 | P62753 | S235 | IAKRRRLSSLRASTS | + |
| S6 | P62753 | S236 | AKRRRLSSLRASTSK | + |
| S6 | P62753 | S240 | RLSSLRASTSKSESS | + |
| SCN2A | Q99250 | S554 | TYEKRFSSPHQSLLS | |
| SCN2A | Q99250 | S573 | LFSPRRNSRASLFSF | |
| SCN2A | Q99250 | S610 | DNDSRRDSLFVPHRH | |
| SCN2A | Q99250 | S623 | RHGERRHSNVSQASR | |
| SCN2A | Q99250 | S686 | EIRKRRSSSYHVSMD | |
| SCN5A | Q14524 | S525 | TSMKPRSSRGSIFTF | |
| SCN5A | Q14524 | S528 | KPRSSRGSIFTFRRR | |
| SDPR | O95810 | S338 | ESFAEGHSEASLASA | |
| SDPR | O95810 | S51 | EEAIRDNSQVNAVTV | |
| SEC14L2 | O76054 | S289 | VQISRGSSHQVEYEI | |
| Separase | Q14674 | S1501 | TDNWRKMSFEILRGS | |
| SERPINA1 | P01009 | T51 | HPTFNKITPNLAEFA | |
| SERPINA3K | P07759 | S316 | ELNLPKFSIASNYRL | |
| SGK1 | O00141 | T369 | DLINKKITPPFNPNV | + |
| SIK | P57059 | S575 | FQEGRRASDTSLTQG | |
| SLC4A4 | Q9Y6R1 | S1026 | KKKKKKGSLDSDNDD | |
| Slob | Q7Z7A4-7 | S54 | EIALQQVSMFFRSEP | |
| SMTNL1 | A8MU46 | S299 | KAPERRVSAPARPRG | |
| SNAP25 | P60880 | T138 | GGFIRRVTNDARENE | |
| SNAPIN | O95295 | S50 | HVHAVRESQVELREQ | |
| Sorcin | P30626 | T178 | DSFRRRDTAQQGVVN | |
| SOX9 | P48436 | S181 | YQPRRRKSVKNGQAE | + |
| SOX9 | P48436 | S64 | EPDLKKESEEDKFPV | ? |
| Spinophilin | Q96SB3 | S100 | LSLPRASSLNENVDH | |
| Spinophilin | Q96SB3 | S94 | SERGVRLSLPRASSL | |
| SPTBN1 | Q01082 | T2159 | NGATEQRTSSKESSP | |
| SPTBN1 | Q01082 | T346 | QQLQAFNTYRTVEKP | |
| Src | P12931 | S17 | DASQRRRSLEPAENV | - |
| Src | P12931 | Y419 | RLIEDNEYTARQGAK | + |
| SREBP1 | P36956 | S338 | IEKRYRSSINDKIIE | |
| StAR | P49675 | S195 | RCAKRRGSTCVLAGM | |
| StAR | P49675 | S277 | HLRKRLESHPASEAR | |
| StAR | P49675 | S56 | INQVRRRSSLLGSRL | |
| StAR | P49675 | S57 | NQVRRRSSLLGSRLE | |
| STEP | P54829 | S221 | GLQERRGSNVSLTLD | |
| STMN1 | P16949 | S16 | KELEKRASGQAFELI | |
| STMN1 | P16949 | S63 | AAEERRKSHEAEVLK | |
| STMN2 | Q93045 | S50 | KQINKRASGQAFELI | |
| STMN2 | Q93045 | S97 | AAEERRKSQEAQVLK | |
| STXBP5 | Q5T5C0 | S723 | GSSSPHNSDDEQKMN | |
| SUN2 | Q9UH99 | S116 | GSESSRASGLVGRKA | |
| SUN2 | Q9UH99 | S12 | SQRLTRYSQGDDDGS | |
| SUN2 | Q9UH99 | S54 | SSNMKRLSPAPQLGP | |
| SYN1 | P17600 | S9 | NYLRRRLSDSNFMAN | |
| SYN2 | Q92777 | S10 | NFLRRRLSDSSFIAN | |
| SYN3 | O14994 | S9 | NFLRRRLSDSSFMAN | |
| syndecan-1 | P18827 | S285 | MKKKDEGSYSLEEPK | |
| SYT12 | Q8IV01 | S97 | GPPSRKGSLSIEDTF | |
| Tau | P10636 | S672 | RVQSKIGSLDNITHV | |
| Tau | P10636 | S725 | DTSPRHLSNVSSTGS | |
| Tau iso3 | P10636-3 | S266 | RVQSKIGSLDNITHV | |
| Tau iso5 (Tau-C) | P10636-5 | S214 | GSRSRTPSLPTPPTR | |
| Tau iso5 (Tau-C) | P10636-5 | S231 | KKVAVVRTPPKSPSS | |
| Tau iso5 (Tau-C) | P10636-5 | T217 | SRTPSLPTPPTREPK | |
| Tau iso8 | P10636-8 | S262 | NVKSKIGSTENLKHQ | |
| Tau iso8 | P10636-8 | S293 | NVQSKCGSKDNIKHV | |
| Tau iso8 | P10636-8 | S305 | KHVPGGGSVQIVYKP | |
| Tau iso8 | P10636-8 | S324 | KVTSKCGSLGNIHHK | |
| Tau iso9 (Tau-F) | P10636-9 | S396 | GAEIVYKSPVVSGDT | |
| Tau iso9 (Tau-F) | P10636-9 | S404 | PVVSGDTSPRHLSNV | |
| Tau iso9 (Tau-F) | P10636-9 | S409 | GTSPRHLSNVSSTGS | |
| Tau iso9 (Tau-F) | P10636-9 | S416 | SNVSSTGSIDMVDSP | |
| TBXA2R | P21731 | S329 | RLSTRPRSLSLQPQL | |
| TBXA2R iso2 | P21731-2 | S330 | LSTRPRRSLTLWPSL | |
| TBXA2R iso2 | P21731-2 | T332 | TRPRRSLTLWPSLEY | |
| TEF-1 | P28347 | S102 | QVLARRKSRDFHSKL | - |
| TGM2 | P21980 | S215 | GRDCSRRSSPVYVGR | ? |
| TGM2 | P21980 | S216 | RDCSRRSSPVYVGRV | ? |
| TH | P07101 | S182 | LSGVRQVSEDVRSPA | |
| TH | P07101 | S71 | RFIGRRQSLIEDARK | + |
| Titin | Q8WZ42 | S4185 | IQQGAKTSLQEEMDS | |
| TNNI3 | P19429 | S149 | TLRRVRISADAMMQA | |
| TNNI3 | P19429 | S165 | LGARAKESLDLRAHL | |
| TNNI3 | P19429 | S22 | PAPIRRRSSNYRAYA | |
| TNNI3 | P19429 | S23 | APIRRRSSNYRAYAT | |
| TNNI3 | P19429 | S38 | EPHAKKKSKISASRK | |
| TNNI3 | P19429 | S41 | AKKKSKISASRKLQL | |
| TNNI3 | P19429 | S43 | KKSKISASRKLQLKT | |
| TNNI3 | P19429 | S76 | GEKGRALSTRCQPLE | |
| TNNI3 | P19429 | T142 | RGKFKRPTLRRVRIS | |
| TP2 | Q05952 | S109 | HRAKRRSSGRRYK__ | |
| TP2 | Q05952 | V125 | MAKRIQQvYKTKTRS | |
| TPH1 | P17752 | S58 | RKSKRRNSEFEIFVD | |
| TPH2 | Q8IWU9 | S104 | RKSRRRSSEVEIFVD | |
| TPH2 | Q8IWU9 | S19 | YWARRGFSLDSAVPE | |
| TPPP | O94811 | S159 | SGVTKAISSPTVSRL | |
| TPPP | O94811 | S32 | DRAAKRLSLESEGAG | |
| TPPP | O94811 | T92 | VIDGRNVTVTDVDIV | |
| TRPV5 | Q9NQA5 | T708 | WEILRQNTLGHLNLG | |
| TTF-1 | P43699 | S336 | DLAHHAASPAALQGQ | |
| TTF-1 | P43699 | T9 | SMSPKHTTPFSVSDI | |
| UHRF1 | Q96T88 | S298 | DNPMRRKSGPSCKHC | |
| VASP | P50552 | S156 | EHIERRVSNAGGPPA | |
| VASP | P50552 | S238 | GAKLRKVSKQEEASG | |
| VASP | P50552 | T277 | LARRRKATQVGEKTP | |
| Vax2 | Q9UIW0 | S170 | RDLEKRASSSASEAF | ? |
| VDR | P11473 | S182 | TPSFSGDSSSSCSDH | - |
| Vimentin | P08670 | S25 | PGTASRPSSSRSYVT | |
| Vimentin | P08670 | S39 | TTSTRTYSLGSALRP | - |
| Vimentin | P08670 | S412 | EGEESRISLPLPNFS | |
| Vimentin | P08670 | S47 | LGSALRPSTSRSLYA | |
| Vimentin | P08670 | S51 | LRPSTSRSLYASSPG | |
| Vimentin | P08670 | S66 | GVYATRSSAVRLRSS | |
| Vimentin | P08670 | S7 | _MSTRSVSSSSYRRM | |
| Vimentin | P08670 | S72 | SSAVRLRSSVPGVRL | |
| Vimentin | P08670 | S73 | SAVRLRSSVPGVRLL | |
| VR1 | Q8NER1 | S117 | LRLYDRRSIFEAVAQ | |
| VR1 | Q8NER1 | S502 | YFLQRRPSMKTLFVD | |
| VR1 | Q8NER1 | S6 | __MEQRASLDSEESE | |
| VR1 | Q8NER1 | S775 | EGVKRTLSFSLRSSR | |
| VR1 | Q8NER1 | S821 | YLRQFSGSLKPEDAE | |
| VR1 | Q8NER1 | T145 | QKSKKHLTDNEFKDP | |
| VR1 | Q8NER1 | T371 | RHLSRKFTEWAYGPV | |
| VTN | P04004 | S397 | NQNSRRPSRATWLSL | |
| WT1 | P19544 | S365 | KDCERRFSRSDQLKR | |
| WT1 | P19544 | S393 | KTCQRKFSRSDHLKT | |
| Zyxin | Q15942 | S142 | PQPREKVSSIDLEID | |
| Zyxin | Q15942 | S143 | QPREKVSSIDLEIDS | |
| 5-LO (ALOX5) | P09917 | S272 | CSLERQLSLEQEVQQ | + |
| MAPT/TAU | P10636 | S579 | NVKSKIGSTENLKHQ | |
| MYLK1 (smMLCK) | Q15746 | S1388 | LATCRSTSFNVQDLL | |
| 14-3-3 eta | Q04917 | S58 | VVGARRSSWRVISSI | |
| 14-3-3 zeta (YWHAZ) | P63104 | S58 | VVGARRSSWRVVSSI | |
| 5-LO | P09917 | S271 | CSLERQLSLEQEVQQ | |
| 5-LO | P09917 | S523 | GMRGRKSSGFPKSVK | - |
| AA-NAT | Q16613 | S205 | HPFLRRNSGC_____ | |
| AA-NAT | Q16613 | T31 | PSCQRRHTLPASEFR | |
| ABCA1 | O95477 | S1042 | GGMQRKLSVALAFVG | |
| ABCA1 | O95477 | S2054 | GGNKRKLSTAMALIG | |
| ABCB1 | P08183 | S667 | SSLIRKRSTRRSVRG | |
| ABCB1 | P08183 | S671 | RKRSTRRSVRGSQAQ | |
| ABCB1 | P08183 | S683 | QAQDRKLSTKEALDE | |
| ABCC9 | O60706 | S1468 | RAFVRKSSILIMDEA | |
| ABCC9 | O60706 | T635 | FESCKKHTGVQPKTI | |
| ACC1 | Q13085 | S1201 | IPTLNRMSFSSNLNH | |
| ACC1 | Q13085 | S78 | LALHIRSSMSGLHLV | |
| ACTA1 | P68133 | S201 | ILTERGYSFVTTAER | |
| ACTA1 | P68133 | S35 | APRAVFPSIVGRPRH | |
| ACTB | P60709 | S33 | APRAVFPSIVGRPRH | |
| ACTG1 | P63261 | S33 | APRAVFPSIVGRPRH | |
| ACTG2 | P63267 | S200 | ILTERGYSFVTTAER | |
| ACTG2 | P63267 | S34 | APRAVFPSIVGRPRH | |
| ACTN1 | P12814 | S404 | EKFRQKASIHEAWTD | |
| ACTN4 | O43707 | S423 | EKFRQKASIHEAWTD | |
| ADD1 | P35611 | S408 | REKSKKYSDVEVPAS | |
| ADD1 | P35611 | S436 | TCSPLRHSFQKQQRE | |
| ADD1 | P35611 | S481 | KEDGHRTSTSAVPNL | |
| ADD1 | P35611 | S59 | MEQKKRVSMILQSPA | |
| ADD1 | P35611 | S726 | KKKFRTPSFLKKSKK | |
| ADD2 | P35612 | S713 | KKKFRTPSFLKKSKK | |
| ADD2 | P35612 | S726 | KKKEKVES_______ | |
| ADD2 | P35612 | T55 | MEQKKRVTMILQSPS | |
| ADD3 | Q9UEY8 | S693 | KKKFRTPSFLKKNKK | |
| ADRB1 | P08588 | S312 | RAGKRRPSRLVALRE | |
| ADRB2 | P07550 | S262 | GHGLRRSSKFCLKEH | |
| ADRB2 | P07550 | S345 | ELLCLRRSSLKAYGN | |
| ADRB2 | P07550 | S346 | LLCLRRSSLKAYGNG | |
| AHNAK | Q09666 | S135 | KIKPRLKSEDGVEGD | |
| AHNAK | Q09666 | S5464 | EGPKVKGSLGATGEI | |
| AHNAK | Q09666 | T158 | ITVTRRVTAYTVDVT | |
| AID | Q9GZX7 | S38 | YVVKRRDSATSFSLD | |
| AID | Q9GZX7 | T27 | WAKGRRETYLCYVVK | |
| AKAP13 | Q12802 | S1565 | LSPFRRHSWGPGKNA | |
| AKAP13 | Q12802 | S2733 | SVSPKRNSISRTHKD | |
| AKAP9 | Q99996 | S55 | QKKKRKTSSSKHDVS | |
| Aldolase A | P04075 | S46 | SIAKRLQSIGTENTE | |
| AMPKa1 (PRKAA1) | Q13131 | S487 | ATPQRSGSVSNYRSC | |
| ANPa R | P16066 | S529 | RHLRSAGSRLTLSGR | + |
| ANXA1 | P04083 | T215 | AGERRKGTDVNVFNT | |
| AP-2 alpha | P05549 | S239 | AEVQRRLSPPECLNA | |
| APOBEC3G | Q9HC16 | T32 | PILSRRNTVWLCYEV | |
| AQP2 | P41181 | S256 | REVRRRQSVELHSPQ | |
| ARFGEF1 | Q9Y6D6 | S883 | EIAGKKISMKETKEL | |
| ARHGDIA | P52565 | S174 | KGMLARGSYSIKSRF | |
| ARHGEF7 (BETAPIX) | Q14155 | S694 | RKPERKPSDEEFASR | + |
| ARHGEF7 iso2 | Q14155-2 | T654 | EFASRKSTAALEEDA | |
| ARPP-21 | Q86V31 | S58 | QERRKSKSGAGKGKL | |
| ASIC1 | P78348 | S524 | CQKEAKRSSADKGVA | |
| ASIC1 | P78348 | S525 | QKEAKRSSADKGVAL | |
| ATF2 | P15336 | S62 | FGPARNDSVIVADQT | + |
| ATP1A1 | P05023 | S943 | ICKTRRNSVFQQGMK | - |
| ATP5J | P18859 | S57 | YKSKRQTSGGPVDAS | |
| AurA (STK15) | O14965 | T288 | APSSRRTTLCGTLDY | + |
| AurC (STK13) | Q9UQB9 | T198 | TPSLRRKTMCGTLDY | + |
| AVPR2 | P30518 | S255 | RRGRRTGSPGEGAHV | |
| B-CK | P12277 | S178 | GRYYALKSMTEAEQQ | |
| B-Raf | P15056 | S429 | PQRERKSSSSSEDRN | - |
| B-Raf | P15056 | S446 | KTLGRRDSSDDWEIP | + |
| Bad | Q92934 | S118 | GRELRRMSDEFVDSF | - |
| Bad | Q92934 | S75 | EIRSRHSSYPAGTED | - |
| Bad | Q92934 | S99 | PFRGRSRSAPPNLWA | - |
| BARK1 (GRK2, ADRBK1) | P25098 | S685 | VPLVQRGSANGL___ | |
| BCAM (Lu) | P50895 | S621 | SGGARGGSGGFGDEC | + |
| bFGF | P09038 | T254 | TYRSRKYTSWYVALK | |
| BIRC5 | O15392 | S20 | FLKDHRISTFKNWPF | |
| BRSK2 | Q8IWQ3 | T260 | VDAARRLTLEHIQKH | + |
| C-EBPb | P17676 | S288 | NNIAVRKSRDKAKMR | |
| CACNA1B | Q00975 | S2129 | SEKQRFYSCDRFGGR | |
| CACNA1C | Q13936 | S1981 | ASLGRRASFHLECLK | |
| CACNA1D | Q01668 | S121 | PIRRACISIVEWKPF | |
| CACNA1D | Q01668 | S1659 | PEIRRAISCDLQDDE | |
| CACNA1D | Q01668 | S1700 | VNSDRRDSLQQTNTT | |
| CACNA1D | Q01668 | S1745 | HNHHNHNSIGKQVPT | |
| CACNA1D | Q01668 | S1816 | AAHGKRPSIGDLEHV | |
| CACNA1D | Q01668 | S1902 | DSPVCYDSRRSPRRR | |
| CACNA1D | Q01668 | S1922 | PASHRRSSFNFECLR | |
| CACNA1D | Q01668 | S2123 | NGDVGPLSHRQDYEL | |
| CACNA1D | Q01668 | S45 | GPTSQPNSSKQTVLS | |
| CACNA1D | Q01668 | S46 | PTSQPNSSKQTVLSW | |
| CACNA1D | Q01668 | S497 | GSLCQAISKSKLSRR | |
| CACNA1D | Q01668 | S499 | LCQAISKSKLSRRWR | |
| CACNA1D | Q01668 | S52 | SSKQTVLSWQAAIDA | |
| CACNA1D | Q01668 | S799 | EVNQIANSDNKVTID | |
| CACNA1D | Q01668 | S864 | IAPIPEGSAFFILSK | |
| CACNA1D | Q01668 | S870 | GSAFFILSKTNPIRV | |
| CACNA1D | Q01668 | T1752 | SIGKQVPTSTNANLN | |
| CACNA1D | Q01668 | T2105 | EMESAASTLLNGNVR | |
| CACNA1D | Q01668 | T443 | KGYLDWITQAEDIDP | |
| CACNA1D | Q01668 | T49 | QPNSSKQTVLSWQAA | |
| CACNA1D | Q01668 | T804 | ANSDNKVTIDDYREE | |
| CACNA1H | O95180 | S1107 | LPDSRRGSSSSGDPP | |
| CACNA1S | Q13698 | S1617 | NFLERTNSLPPVMAN | |
| CACNA1S | Q13698 | S687 | EKKRRKMSKGLPDKS | |
| CACNB2 | Q08289 | S514 | SAPIRSASQAEEEPS | |
| CACNB2 | Q08289 | S533 | KKSQHRSSSSAPHHN | |
| CACNB2 | Q08289 | S534 | KSQHRSSSSAPHHNH | |
| CACNG2 | Q9Y698 | T321 | NTANRRTTPV_____ | |
| CAD | P27708 | S1406 | GAGGRRLSSFVTKGY | + |
| CAD | P27708 | S1859 | PPRIHRASDPGLPAE | + |
| Calnexin (CANX) | P27824 | S583 | EDEILNRSPRNRKPR | |
| Calpain 2 | P17655 | S368 | DGNWRRGSTAGGCRN | |
| Calpain 2 | P17655 | T369 | GNWRRGSTAGGCRNY | |
| CaMKK1 | Q8N5S9 | S458 | KSMLRKRSFGNPFEP | - |
| CaMKK1 | Q8N5S9 | S475 | RREERSMSAPGNLLV | - |
| CaMKK1 | Q8N5S9 | S52 | PPRARAASVIPGSTS | |
| CaMKK1 | Q8N5S9 | S74 | SLSARKLSLQERPAG | + |
| CaMKK1 | Q8N5S9 | T108 | PRAWRRPTIESHHVA | + |
| Caspase 9 | P55211 | S183 | GLRTRTGSNIDCEKL | |
| Caspase 9 | P55211 | S195 | EKLRRRFSSLHFMVE | |
| Caspase 9 | P55211 | S99 | NRQAAKLSKPTLENL | |
| CBX3 | Q13185 | S93 | KDGTKRKSLSDSESD | |
| CD44 | P16070 | S697 | AVEDRKPSGLNGEAS | |
| Ces3 | Q6UWW8 | S506 | ANFARNGSPNGGGLP | |
| CETN2 | P41208 | S170 | LRIMKKTSLY_____ | |
| CFTR | P13569 | S660 | FSAERRNSILTETLH | |
| CFTR | P13569 | S700 | FGEKRKNSILNPINS | |
| CFTR | P13569 | S712 | INSIRKFSIVQKTPL | |
| CFTR | P13569 | S737 | EPLERRLSLVPDSEQ | |
| CFTR | P13569 | S753 | EAILPRISVISTGPT | |
| CFTR | P13569 | S768 | LQARRRQSVLNLMTH | |
| CFTR | P13569 | S795 | TASTRKVSLAPQANL | |
| CFTR | P13569 | S813 | DIYSRRLSQETGLEI | |
| CGb | P01233 | T117 | CALCRRSTTDCGGPK | |
| CHCHD3 | Q9NX63 | T11 | TTSTRRVTFEADENE | |
| CIITA | P33076 | S1050 | AASLLRLSLYNNCIC | |
| CIITA | P33076 | S834 | QELPGRLSFLGTRLT | |
| CK1d (CSNK1D) | P48730 | S370 | MERERKVSMRLHRGA | - |
| Claudin 3 | O15551 | T192 | PPREKKYTATKVVYS | |
| COL4A3 | Q01955 | S1435 | LKGKRGDSGSPATWT | |
| COL4A3 | Q01955 | S1452 | GFVFTRHSQTTAIPS | |
| Connexin 32 | P08034 | S233 | NPPSRKGSGFGHRLS | |
| Coronin 1C | Q9ULV4 | S354 | IMTVPRKSDLFQDDL | |
| CP | P00450 | T688 | SKGERRDTANLFPHK | |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| CREM | Q03060-2 | S71 | EILSRRPSYRKILNE | |
| CRF-R1 | P34998 | S330 | LMTKLRASTTSETIQ | |
| CSF2RB | P32927 | S601 | LGPPHSRSLPDILGQ | |
| CSK | P41240 | S364 | ALREKKFSTKSDVWS | + |
| CSP | Q9H3Z4 | S10 | DQRQRSLSTSGESLY | - |
| CTNNB1 | P35222 | S45 | GATTTAPSLSGKGNP | + |
| CTNNB1 | P35222 | S552 | QDTQRRTSMGGTQQQ | |
| CTNNB1 | P35222 | S675 | QDYKKRLSVELTSSL | |
| CTPS | P17812 | T455 | MRLGKRRTLFQTKNS | |
| CUX1 (CUTL1) | P39880 | S1215 | AYMKRRHSSVSDSQP | - |
| CUX1 (CUTL1) | P39880 | S1237 | TEYSQGASPQPQHQL | - |
| CUX1 (CUTL1) | P39880 | S1270 | YQQKPYPSPKTIEDL | - |
| Cx36 | Q9UKL4 | S110 | KQRERRYSTVFLALD | |
| Cx36 | Q9UKL4 | S293 | GAQAKRKSIYEIRNK | |
| Cx36 | Q9UKL4 | S306 | NKDLPRVSVPNFGRT | |
| Cx36 | Q9UKL4 | S315 | PNFGRTQSSDSAYV_ | |
| Cx43 | P17302 | S364 | IVDQRPSSRASSRAS | |
| Cx43 | P17302 | S367 | QRPSSRASSRASSRP | |
| Cx43 | P17302 | S368 | RPSSRASSRASSRPR | |
| Cx43 | P17302 | S372 | RASSRASSRPRPDDL | |
| Cyclin D1 (CCND1) | P24385 | S197 | ATDVKFISNPPSMVA | |
| Cyclin D1 (CCND1) | P24385 | S234 | YRLTRFLSRVIKCDP | |
| Cyclin D1 (CCND1) | P24385 | S90 | NYLDRFLSLEPVKKS | |
| CYP3A5 | P20815 | S392 | GVFIPKGSMVVIPTY | |
| DAP3 | P51398 | S185 | FDQPLEASTWLKNFK | |
| DAP3 | P51398 | S220 | RESTEKGSPLGEVVE | |
| DAP3 | P51398 | S31 | MGTQARQSIAAHLDN | |
| DAP3 | P51398 | T186 | DQPLEASTWLKNFKT | |
| DARPP-32 | Q9UD71 | S137 | EEEEEEDSQAEVLKV | |
| DARPP-32 | Q9UD71 | T34 | MIRRRRPTPAMLFRL | |
| Dematin | Q08495 | S403 | NELKKKASLF_____ | |
| Desmin | P17661 | S11 | YSSSQRVSSYRRTFG |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
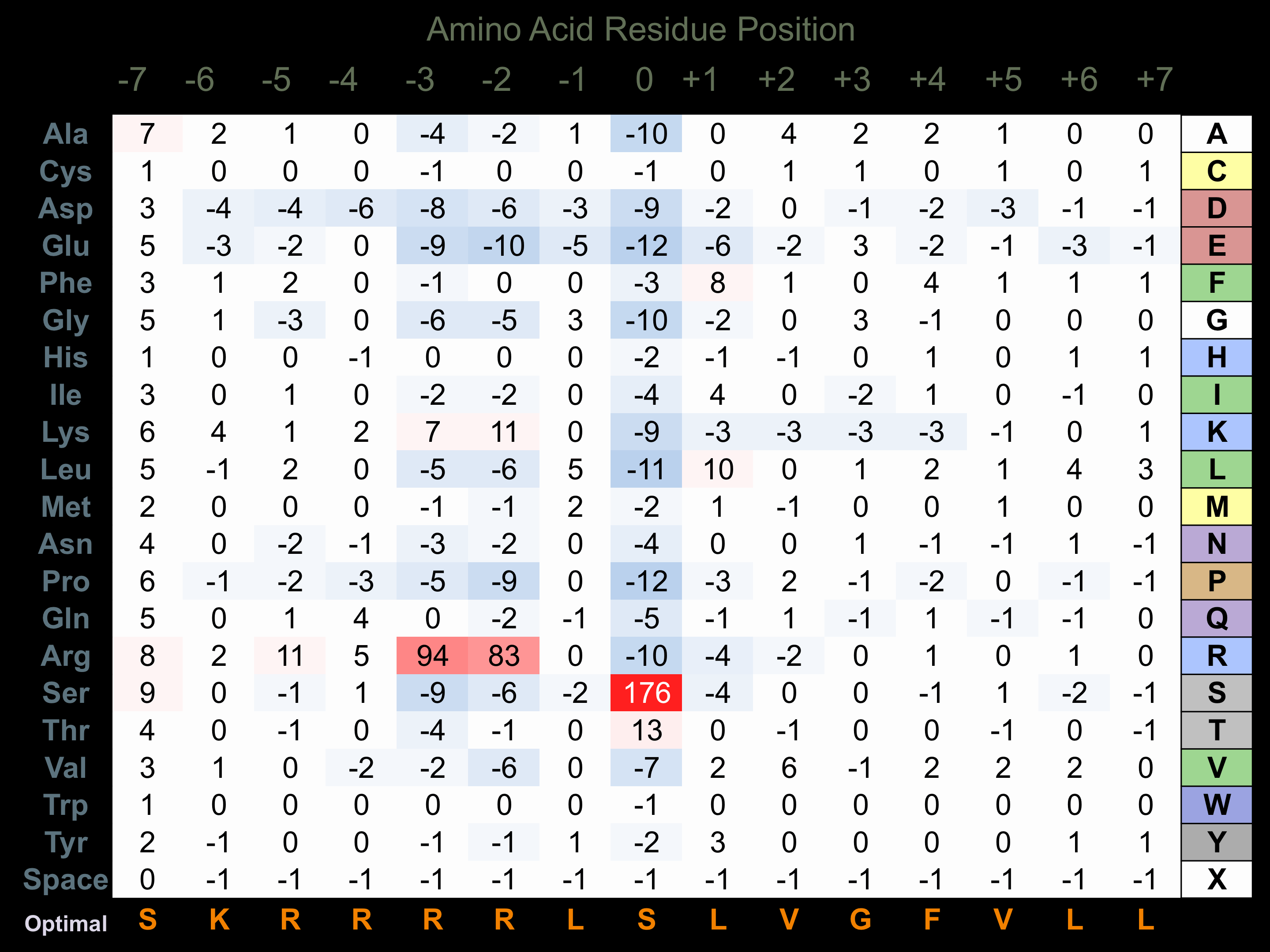
Matrix Type:
Experimentally derived from alignment of 1115 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, endocrine disorders
Specific Diseases (Non-cancerous):
Primary pigmented nodular adrenocortical disease (PPNAD); Cushing syndrome, ACTH-independent adrenal, somatic
Specific Cancer Types:
Hepatocellular fibrolamellar carcinomas
Comments:
PKACa may be an oncoprotein (OP), although this kinase has also been implicated in inhibiting normal cell proliferation. Gain-of-function mutations in the PRKACA catalytic domain are associated with elevated catalytic activity of the kinase, and have been observed in several cancer types. In particular, a L206R subsitution mutation is observed in cancer and in Cushing's syndrome patients. The gain-of-function mutation is predicted to result in the interruption of the interaction between the catalytic and regulatory subunits of protein kinase A, resulting in a constitutively active PKA. Gain-of-function mutations in the PRKACA gene that lead to consitutive activity of the PKA holoenzyme have been suggested to play an important role in the development of several types of cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +880, p<(0.0003); Brain oligodendrogliomas (%CFC= +968, p<0.076); and Uterine leiomyomas (%CFC= +182, p<0.003). The COSMIC website notes an up-regulated expression score for PKACa in diverse human cancers of 466, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 91 for this protein kinase in human cancers was 1.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25371 diverse cancer specimens. This rate is only 18 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.39 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: L206R (4).
Comments:
Only 2 deletions and 1 insertion, and no complex mutations are noted on the COSMIC website.

