Nomenclature
Short Name:
CDK9
Full Name:
Cyclin-dependent kinase 9
Alias:
- C-2K
- CDC2L4
- Kinase Cdk9
- PITALRE
- TAK
- Cell division cycle 2-like protein kinase 4
- Cyclin-dependent kinase 9
- EC 2.7.11.22
- EC 2.7.11.23
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
CDK9
Specific Links
Structure
Mol. Mass (Da):
42,778
# Amino Acids:
372
# mRNA Isoforms:
2
mRNA Isoforms:
53,365 Da (489 AA; P50750-2); 42,778 Da (372 AA; P50750)
4D Structure:
Associates with CCNT1/cyclin-T1 to form P-TEFb. P-TEFb forms a complex with AFF4/AF5Q31. Also associates with CCNK/cyclin-K. Component of a complex which is at least composed of HTATSF1/Tat-SF1, P-TEFb complex, RNA pol II, SUPT5H, and NCL/nucleolin. Component of the 7SK snRNP complex at least composed of P-TEFb (composed of CDK9 and CCNT1/cyclin-T1), HEXIM1, HEXIM2, BCDIN3, SART3 proteins and 7SK and U6 snRNAs.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 19 | 315 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K35, K44, K48, K49, K56, K68, K74, K127, K164, K178, K269, K274, K276, K345.
Methylated:
K21, K35, R86, K127, R370.
Serine phosphorylated:
S7, S175+, S180+, S317, S334, S347+, S353+, S357+.
Threonine phosphorylated:
T186+, T330, T333, T350+, T354+, T362, T363, T366.
Tyrosine phosphorylated:
Y185+, Y194-, Y338.
Ubiquitinated:
K24, K35, K56, K68, K88, K151, K164, K178, K269, K294, K345.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 48
48
911
31
927
 9
9
170
12
193
 6
6
123
7
131
 9
9
179
104
331
 33
33
631
32
509
 100
100
1895
67
5475
 10
10
182
39
419
 26
26
489
33
967
 21
21
391
10
241
 3
3
65
95
79
 8
8
153
22
132
 34
34
647
114
596
 9
9
168
18
209
 13
13
239
9
212
 11
11
207
19
202
 3
3
60
18
59
 5
5
90
192
167
 6
6
113
14
121
 3
3
57
91
44
 23
23
444
112
400
 13
13
248
18
195
 14
14
258
20
203
 9
9
168
16
191
 11
11
216
14
200
 17
17
316
18
331
 28
28
530
69
555
 8
8
145
21
125
 9
9
176
14
143
 13
13
253
14
207
 6
6
112
42
106
 62
62
1167
18
796
 38
38
717
30
1762
 2
2
32
77
86
 36
36
678
83
596
 11
11
214
48
415
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 92.4
92.4
93.5
- -
-
-
99 -
-
-
99 68.5
68.5
69.4
98 -
-
-
- 98.6
98.6
99.7
99 98.6
98.6
99.7
99 -
-
-
- 77.9
77.9
80.3
- 93
93
95.4
93 89.3
89.3
94.1
91 85.5
85.5
89.5
91 -
-
-
- 66
66
75.9
74 72.7
72.7
83.2
- 43.3
43.3
56.4
57 67.3
67.3
75.8
- -
-
-
- -
-
-
- -
-
-
- 36
36
51
- 25.2
25.2
39.4
41 31.7
31.7
45.6
34
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CCNT1 - O60563 |
| 2 | POLR2A - P24928 |
| 3 | CCNT2 - O60583 |
| 4 | SKP2 - Q13309 |
| 5 | CUL1 - Q13616 |
| 6 | TP53 - P04637 |
| 7 | RB1 - P06400 |
| 8 | NBN - O60934 |
| 9 | NFKB1 - P19838 |
| 10 | STK36 - Q9NRP7 |
| 11 | MED21 - Q13503 |
| 12 | HEXIM2 - Q96MH2 |
| 13 | RELA - Q04206 |
| 14 | CDC34 - P49427 |
| 15 | SKP1 - P63208 |
Regulation
Activation:
Phosphorylation at Ser-175 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
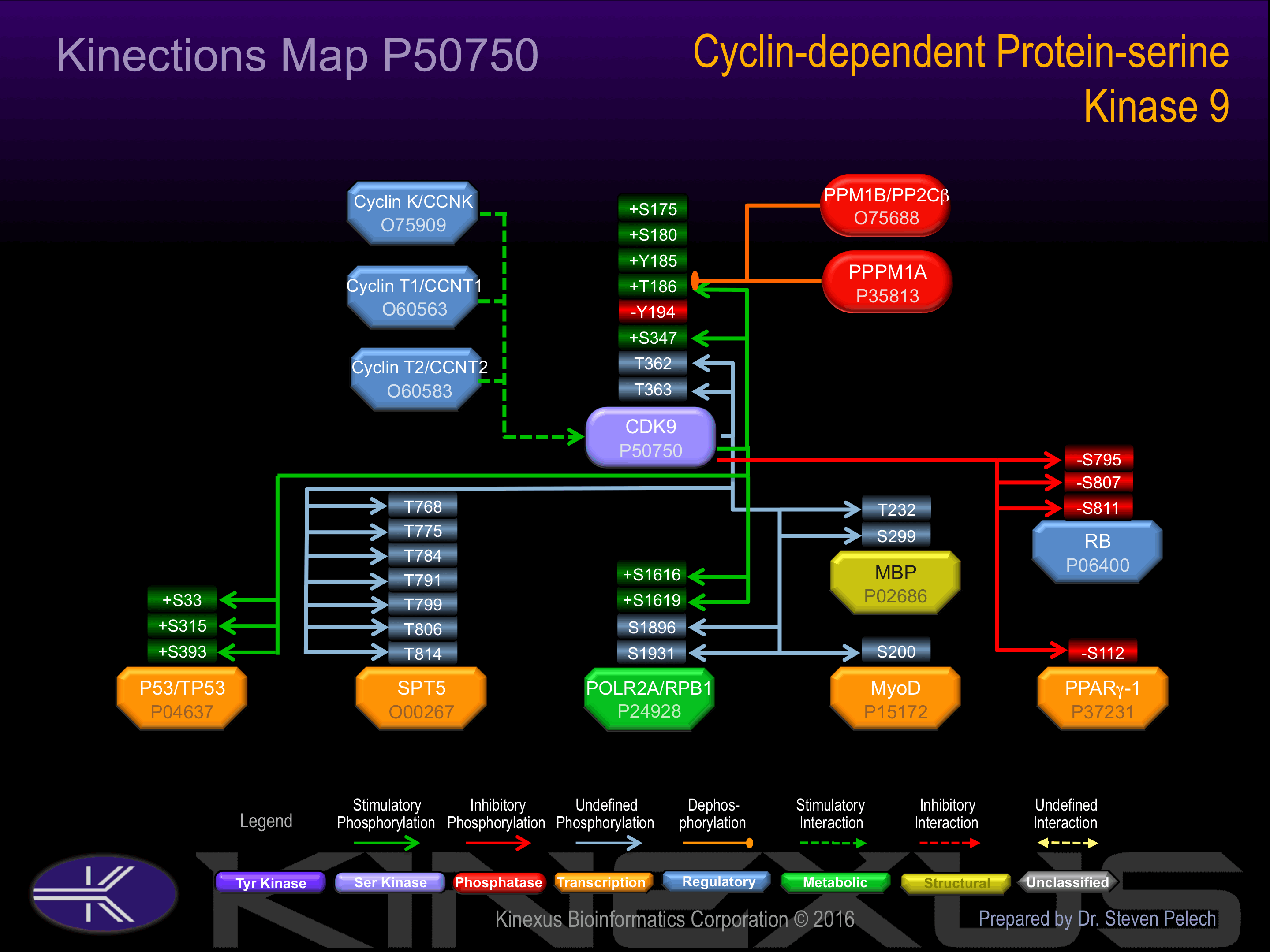
| CDK9 | P50750 | S347 | APPRRKGSQITQQST | + |
| CDK9 | P50750 | T186 | NSQPNRYTNRVVTLW | + |
| CDK9 | P50750 | T362 | NQSRNPATTNQTEFE | |
| CDK9 | P50750 | T363 | QSRNPATTNQTEFER | |
| MBP | P02686 | S299 | GRDSRSGSPMARR__ | |
| MBP | P02686 | T232 | KNIVTPRTPPPSQGK | |
| MyoD | P15172 | S200 | SGDSDASSPRSNCSD | |
| p53 | P04637 | S315 | LPNNTSSSPQPKKKP | + |
| p53 | P04637 | S33 | LPENNVLSPLPSQAM | + |
| p53 | P04637 | S392 | FKTEGPDSD______ | + |
| POLR2A (RPB1) | P24928 | S1616 | TPQSPSYSPTSPSYS | + |
| POLR2A (RPB1) | P24928 | S1619 | SPSYSPTSPSYSPTS | + |
| POLR2A (RPB1) | P24928 | S1896 | SPTSPTYSPTSPVYT | |
| POLR2A (RPB1) | P24928 | S1931 | SPTSPTYSPTSPKGS | |
| PPARg-1 | P37231 | S112 | AIKVEPASPPYYSEK | - |
| Rb | P06400 | S795 | SPYKFPSSPLRIPGG | - |
| Rb | P06400 | S807 | PGGNIYISPLKSPYK | - |
| Rb | P06400 | S811 | IYISPLKSPYKISEG | - |
| SPT5 | O00267 | T768 | MTSTYGRTPMYGSQT | |
| SPT5 | O00267 | T775 | TPMYGSQTPMYGSGS | |
| SPT5 | O00267 | T784 | MYGSGSRTPMYGSQT | |
| SPT5 | O00267 | T791 | TPMYGSQTPLQDGSR | |
| SPT5 | O00267 | T799 | PLQDGSRTPHYGSQT | |
| SPT5 | O00267 | T806 | TPHYGSQTPLHDGSR | |
| SPT5 | O00267 | T814 | PLHDGSRTPAQSGAW |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
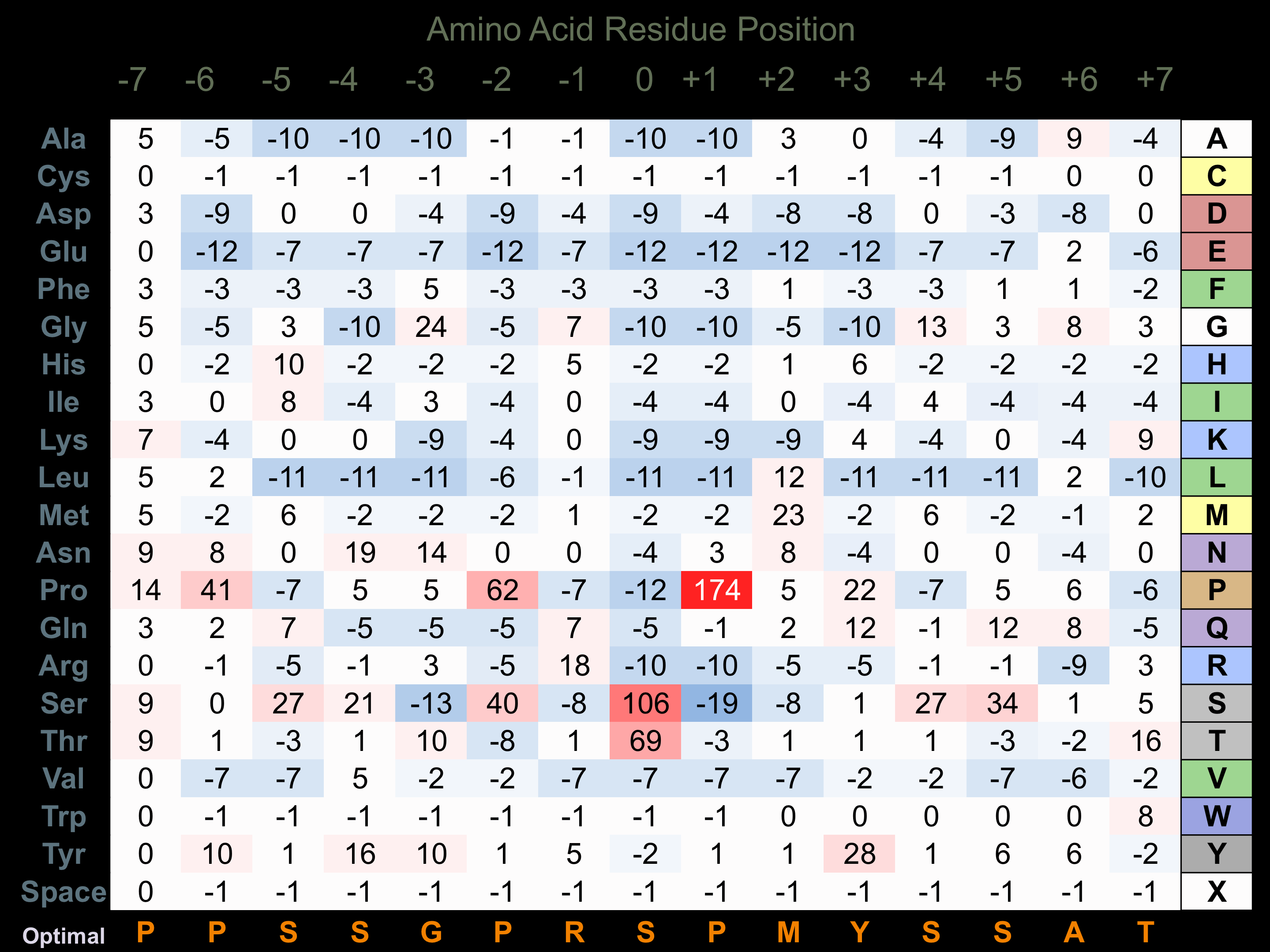
Matrix Type:
Experimentally derived from alignment of 33 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Infectious disease
Specific Diseases (Non-cancerous):
Hiv-1
Comments:
Chronic activation of CDK9 enlarges cardiac myocyte, which may result in cardiac hypertrophy, and predisposition to heart failure. Mutations on 347-357 positions lead to loss of autophosphorylation and can either impair or promote interaction with HIV TAT.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Malignant pleural mesotheliomas (MPM) tumours (%CFC= +126, p<0.004); Prostate cancer - primary (%CFC= -46, p<0.0001); Skin melanomas - malignant (%CFC= +428, p<0.0001); and Uterine leiomyomas (%CFC= +61, p<0.084).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24433 diverse cancer specimens. This rate is only -22 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.32 % in 1270 large intestine cancers tested; 0.31 % in 864 skin cancers tested.
Frequency of Mutated Sites:
None > 1 in 20,197 cancer specimens
Comments:
Only 3 insertions (I210fs*2 insertions frameshift), and no deletions or complex mutations are noted on the COSMIC website.