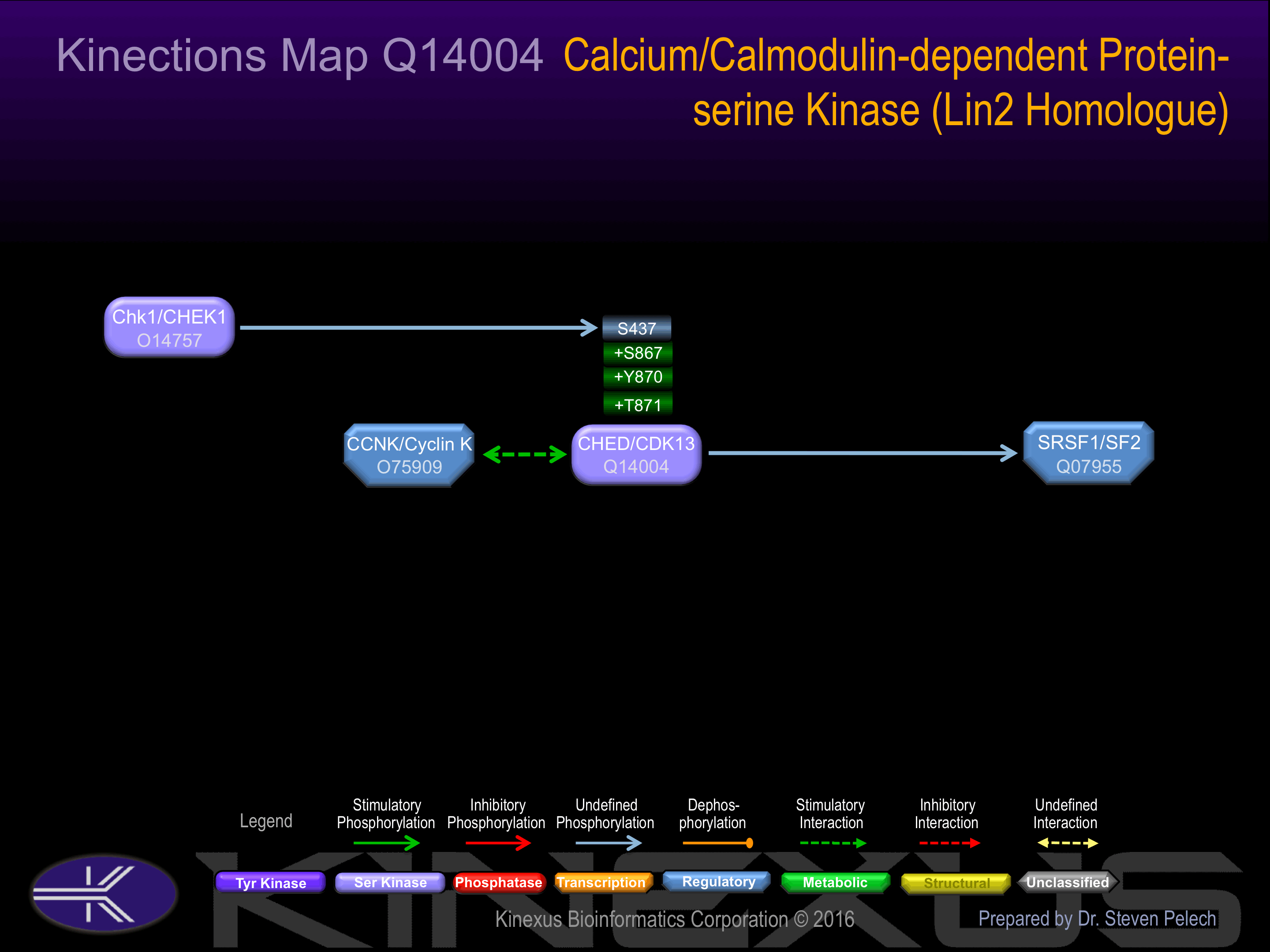
Nomenclature
Short Name:
CHED
Full Name:
Cholinesterase-related cell division controller
Alias:
- CD2L5
- CDC2L5
- CDC2-related protein kinase 5
- CDK13
- Cell division cycle 2-like 5
- EC 2.7.11.22
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
CRK7
Specific Links
Structure
Mol. Mass (Da):
164,923
# Amino Acids:
1512
# mRNA Isoforms:
2
mRNA Isoforms:
164,923 Da (1512 AA; Q14004); 158,435 Da (1452 AA; Q14004-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
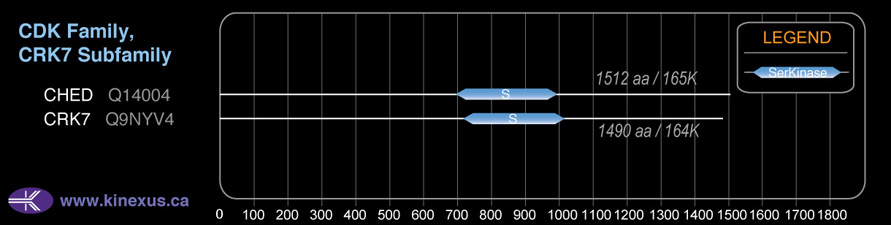
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 705 | 998 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K556, K566, K583, K585, K951.
Serine phosphorylated:
S3, S4, S5, S135, S136, S151, S204, S205, S209, S229, S238, S260, S262, S266, S268, S270, S276, S281, S285, S291, S315, S317, S325, S328, S337, S340, S342, S348, S349, S352, S358, S360, S363, S367, S369, S371, S374, S375, S383, S385, S391, S395, S397, S400, S409, S411, S413, S431, S433, S436, S437, S439, S441, S490, S493, S507, S525, S527, S528, S590, S624, S630, S664+, S867+, S1022, S1048, S1054, S1066, S1146, S1153, S1163, S1225, S1229, S1342, S1346.
Threonine phosphorylated:
T141, T264, T442, T444, T494, T496, T500, T502, T531, T539, T588, T748, T871+, T1034, T1056, T1058, T1147, T1226, T1246.
Tyrosine phosphorylated:
Y351, Y362, Y370, Y376, Y387, Y399, Y687, Y716+, Y870+, Y1277.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 86
86
994
83
1131
 5
5
57
38
88
 21
21
247
31
204
 37
37
423
288
771
 72
72
827
75
675
 15
15
173
223
576
 26
26
298
99
498
 100
100
1152
116
2543
 39
39
450
38
390
 13
13
145
279
245
 10
10
119
86
146
 48
48
551
461
581
 13
13
151
86
216
 4
4
48
30
38
 11
11
131
70
170
 5
5
59
49
76
 6
6
74
512
146
 16
16
186
52
253
 10
10
111
259
151
 53
53
605
324
693
 10
10
118
65
135
 17
17
201
76
266
 18
18
213
44
189
 26
26
301
51
334
 19
19
217
65
259
 75
75
868
180
1065
 14
14
156
89
221
 11
11
127
52
137
 17
17
195
52
229
 7
7
77
98
54
 45
45
521
42
701
 81
81
929
91
1625
 18
18
213
178
437
 72
72
835
187
721
 71
71
816
109
1913
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.8
100 41.5
41.5
53.5
99 -
-
-
97 -
-
-
94 82.5
82.5
84.6
- -
-
-
- 95.2
95.2
96.9
96 41.7
41.7
53.9
95 -
-
-
- -
-
-
- 29.8
29.8
38
82 -
-
-
68 44
44
55
- -
-
-
- -
-
-
40 -
-
-
- 24.6
24.6
33.3
39 32.6
32.6
47
- -
-
-
- -
-
-
- -
-
-
52 -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PRPF40A - O75400 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
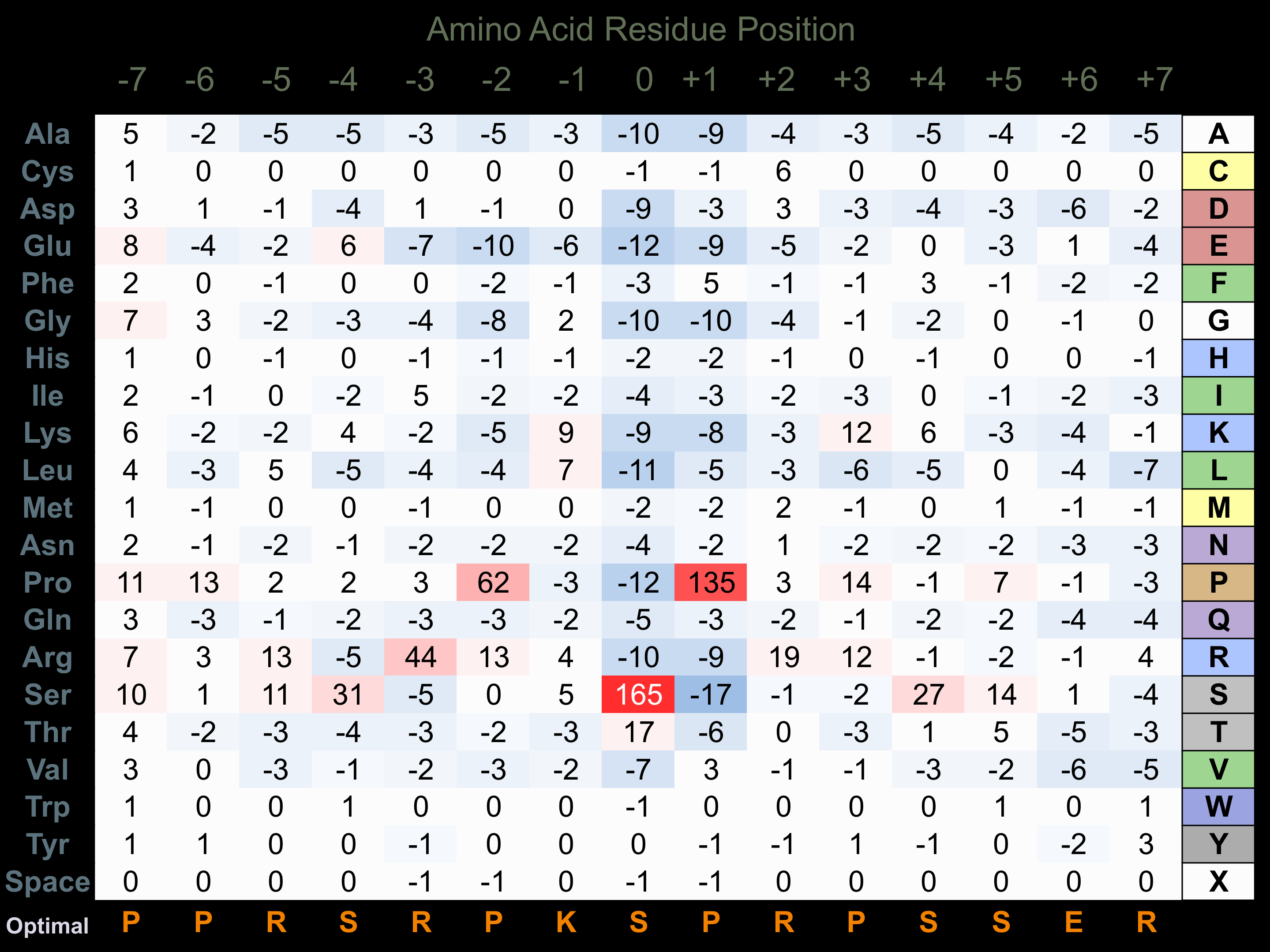
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AST-487 | Kd = 940 pM | 11409972 | 574738 | 22037378 |
| AT7519 | Kd = 3.2 nM | 11338033 | 22037378 | |
| AC1NS7CD | Kd = 23 nM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 23 nM | 3025986 | 296468 | 22037378 |
| Linifanib | Kd = 38 nM | 11485656 | 223360 | 22037378 |
| Staurosporine | Kd = 70 nM | 5279 | 22037378 | |
| R547 | Kd = 110 nM | 6918852 | 22037378 | |
| Foretinib | Kd = 210 nM | 42642645 | 1230609 | 22037378 |
| Alvocidib | Kd = 430 nM | 9910986 | 428690 | 22037378 |
| BMS-345541 | Kd = 800 nM | 9813758 | 249697 | 22037378 |
| Lestaurtinib | Kd = 1 µM | 126565 | 22037378 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Barasertib | Kd = 1.1 µM | 16007391 | 215152 | 22037378 |
| Doramapimod | Kd = 1.1 µM | 156422 | 103667 | 22037378 |
| A674563 | Kd = 3.1 µM | 11314340 | 379218 | 22037378 |
Disease Linkage
General Disease Association:
Cornea, and blood vessel disorders
Specific Diseases (Non-cancerous):
Corneal endothelial dystrophy 2, autosomal recessive; Congenital hereditary endothelial dystrophy Type 1
Comments:
Corneal Endothelial Dystrophy 2, Autosomal Recessive is a heritable recessive disorder involving endotheliitis (inflammation of blood vessels) and corneal dystrophy (deposition of substances in cornea). Congenital Hereditary Endothelial Dystrophy Type I is a dominant inherited disorder involving endotheliitis (inflammation of blood vessels).
Comments:
CHED levels are up-regulated 1.7-fold in human tumours compared to most other protein kinases.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain oligodendrogliomas (%CFC= -57, p<0.098); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +728, p<0.003); and Prostate cancer - primary (%CFC= -61, p<0.0001). The COSMIC website notes an up-regulated expression score for CHED in diverse human cancers of 801, which is 1.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 234 for this protein kinase in human cancers was 3.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24939 diverse cancer specimens. This rate is only -29 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.66 % in 10 peritoneum cancers tested; 0.21 % in 1270 large intestine cancers tested; 0.18 % in 864 skin cancers tested; 0.18 % in 589 stomach cancers tested; 0.15 % in 603 endometrium cancers tested; 0.12 % in 273 cervix cancers tested; 0.09 % in 710 oesophagus cancers tested; 0.08 % in 1634 lung cancers tested; 0.05 % in 1512 liver cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,224 cancer specimens
Comments:
Ten deletions, no insertions and 1 complex mutation are noted on the COSMIC website.