Nomenclature
Short Name:
CHK1
Full Name:
Check point kinase 1
Alias:
- CHEK1
- EC 2.7.11.1
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMKL
SubFamily:
CHK1
Specific Links
Structure
Mol. Mass (Da):
54,420
# Amino Acids:
476
# mRNA Isoforms:
3
mRNA Isoforms:
54,434 Da (476 AA; O14757); 50,415 Da (442 AA; O14757-3); 43,703 Da (382 AA; O14757-2)
4D Structure:
Interacts with BRCA1, CLSPN, FBXO6, PPM1D, RAD51, TIMELESS, XPO1/CRM1 and YWHAZ/14-3-3 zeta.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 9 | 265 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K452.
Serine phosphorylated:
S280-, S284, S286+, S288, S291, S296+, S301, S305, S307, S308, S315, S316, S317+, S331, S333, S343, S345+, S357, S365, S366, S389, S467, S468.
Threonine phosphorylated:
T279, T330, T348, T362.
Ubiquitinated:
K132, K145, K292, K313, K383, K399, K436.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 48
48
1430
35
1562
 0.6
0.6
17
14
18
 2
2
46
12
30
 28
28
846
119
2880
 16
16
489
35
380
 1
1
30
88
29
 3
3
86
41
197
 100
100
2974
43
7103
 13
13
379
17
312
 1.5
1.5
44
96
46
 0.8
0.8
23
33
19
 13
13
393
170
392
 0.9
0.9
28
35
24
 0.6
0.6
19
9
16
 0.8
0.8
25
26
30
 0.3
0.3
10
19
7
 15
15
448
187
3213
 0.7
0.7
21
22
22
 0.9
0.9
28
106
32
 12
12
369
137
363
 1
1
31
23
29
 0.8
0.8
25
27
19
 2
2
49
15
32
 2
2
69
22
68
 3
3
79
24
69
 74
74
2206
75
5976
 3
3
80
38
77
 1.3
1.3
39
19
54
 0.9
0.9
27
20
21
 5
5
150
42
142
 14
14
404
24
398
 59
59
1754
37
6099
 28
28
822
84
1122
 26
26
764
78
691
 5
5
140
48
111
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
100
100 99.4
99.4
99.8
99 -
-
-
96 -
-
-
94 93.3
93.3
95.2
97 -
-
-
- 93.3
93.3
97.1
93 94.3
94.3
97.3
94 -
-
-
- 89.9
89.9
95.4
- 84.5
84.5
91.6
85 77.7
77.7
86.8
78 56.9
56.9
71
65 -
-
-
- 46.1
46.1
62.9
50 45.6
45.6
63.9
- 32
32
48.7
41 58
58
73.3
- -
-
-
- -
-
-
- -
-
-
- 33
33
52.1
- -
-
-
34 -
-
-
37
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC25A - P30304 |
| 2 | TP53 - P04637 |
| 3 | CDC25C - P30307 |
| 4 | CLSPN - Q9HAW4 |
| 5 | CSNK2A1 - P68400 |
| 6 | MDM4 - O15151 |
| 7 | CUL1 - Q13616 |
| 8 | PRKDC - P78527 |
| 9 | XIAP - P98170 |
| 10 | TLK1 - Q9UKI8 |
| 11 | BRCA1 - P38398 |
| 12 | MSH2 - P43246 |
| 13 | AKT1 - P31749 |
| 14 | CSNK2B - P67870 |
| 15 | LY6G5B - Q8NDX9 |
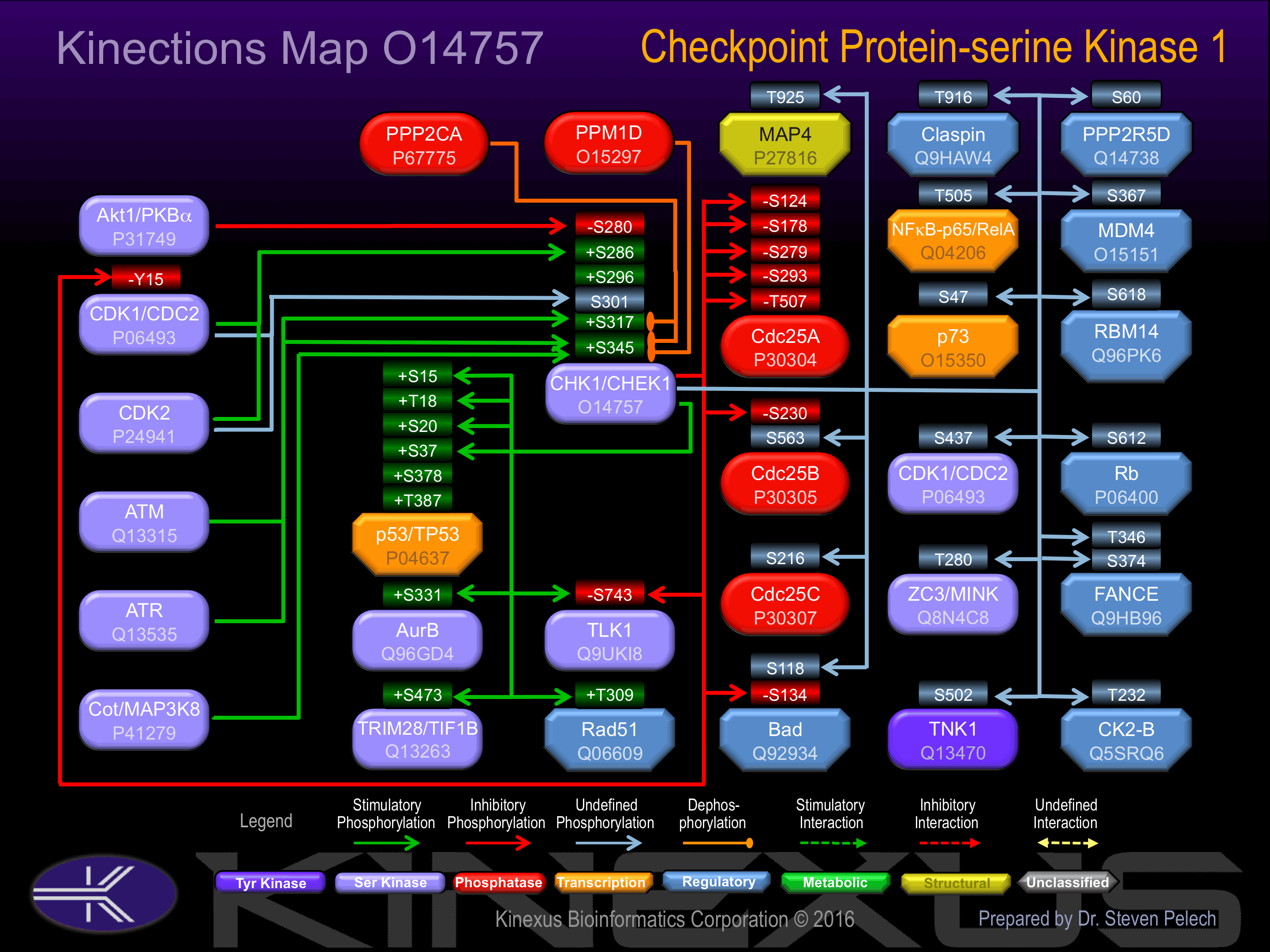
Regulation
Activation:
Activated by phosphorylation at Ser-296, Ser-317, and Ser-345.
Inhibition:
Inhibited by phosphorylation at Ser-280.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Akt1 | P31749 | S280 | AKRPRVTSGGVSESP | - |
| CDK1 | P06493 | S286 | TSGGVSESPSGFSKH | + |
| CDK2 | P24941 | S286 | TSGGVSESPSGFSKH | + |
| CDK1 | P06493 | S301 | IQSNLDFSPVNSASS | ? |
| CDK2 | P24941 | S301 | IQSNLDFSPVNSASS | ? |
| ATR | Q13535 | S317 | ENVKYSSSQPEPRTG | + |
| ATM | Q13315 | S317 | ENVKYSSSQPEPRTG | + |
| ATR | Q13535 | S345 | LVQGISFSQPTCPDH | + |
| ATM | Q13315 | S345 | LVQGISFSQPTCPDH | + |
| COT | P41279 | S345 | LVQGISFSQPTCPDH | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AurB | Q96GD4 | S331 | HPWVRANSRRVLPPS | + |
| Bad | Q92934 | S118 | GRELRRMSDEFVDSF | - |
| Bad | Q92934 | S134 | KGLPRPKSAGTATQM | - |
| Cdc25A | P30304 | S124 | PALKRSHSDSLDHDI | - |
| Cdc25A | P30304 | S178 | LFTQRQNSAPARMLS | - |
| Cdc25A | P30304 | S279 | VLKRPERSQEESPPG | - |
| Cdc25A | P30304 | S293 | GSTKRRKSMSGASPK | - |
| Cdc25A | P30304 | S76 | SNLQRMGSSESTDSG | - |
| Cdc25A | P30304 | T507 | KFRTKSRTWAGEKSK | - |
| Cdc25B | P30305 | S230 | AFAQRPSSAPDLMCL | - |
| Cdc25B | P30305 | S563 | TFRLKTRSWAGERSR | |
| Cdc25C | P30307 | S216 | SGLYRSPSMPENLNR | ? |
| CDK1 (CDC2) | P06493 | Y15 | EKIGEGTYGVVYKGR | - |
| CHED (CDC2L5, CDK13) | Q14004 | S437 | RSRSRHSSISPSTLT | |
| CK2-B | Q5SRQ6 | T232 | NFKSPVKTIR_____ | |
| Claspin | Q9HAW4 | T916 | DELLDLCTGKFTSQA | |
| FANCE | Q9HB96 | S374 | LFLGRILSLTSSASR | |
| FANCE | Q9HB96 | T346 | LGLLRLCTWLLALSP | |
| MAP4 (DKFZp779A1753) | P27816 | T925 | PRLSRLATNTSAPDL | |
| MDM4 | O15151 | S367 | PDCRRTISAPVVRPK | |
| MST1 (STK4) | Q13043 | T340 | AVGDEMGTVRVASTM | |
| NFkB-p105 | P19838 | S328 | INITKPASVFVQLRR | - |
| NFkB-p65 | Q04206 | T505 | EAITRLVTGAQRPPD | |
| p53 | P04637 | S15 | PSVEPPLSQETFSDL | + |
| p53 | P04637 | S20 | PLSQETFSDLWKLLP | + |
| p53 | P04637 | S37 | NVLSPLPSQAMDDLM | + |
| p53 | P04637 | S378 | SKKGQSTSRHKKLMF | + |
| p53 | P04637 | T18 | EPPLSQETFSDLWKL | + |
| p53 | P04637 | T387 | HKKLMFKTEGPDSD_ | + |
| p73 | O15350 | S47 | EVVGGTDSSMDVFHL | |
| PPP2R5D | Q14738 | S60 | PSSNKRPSNSTPPPT | |
| Rad51 | Q06609 | T309 | LRKGRGETRICKIYD | + |
| Rb | P06400 | S612 | MYLSPVRSPKKKGST | - |
| RBM14 | Q96PK6 | S618 | LSDYRRLSESQLSFR | |
| TLK1 | Q9UKI8 | S743 | PHMRRSNSSGNLHMA | - |
| TNK1 | Q13470 | S502 | RMKGISRSLESVLSL | |
| TRIM28 (TIF1B) | Q13263 | S473 | SGVkRSRSGEGEVSG | + |
| ZC3 (MINK) | Q8N4C8 | T280 | TYLSRPPTEQLLKFP |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
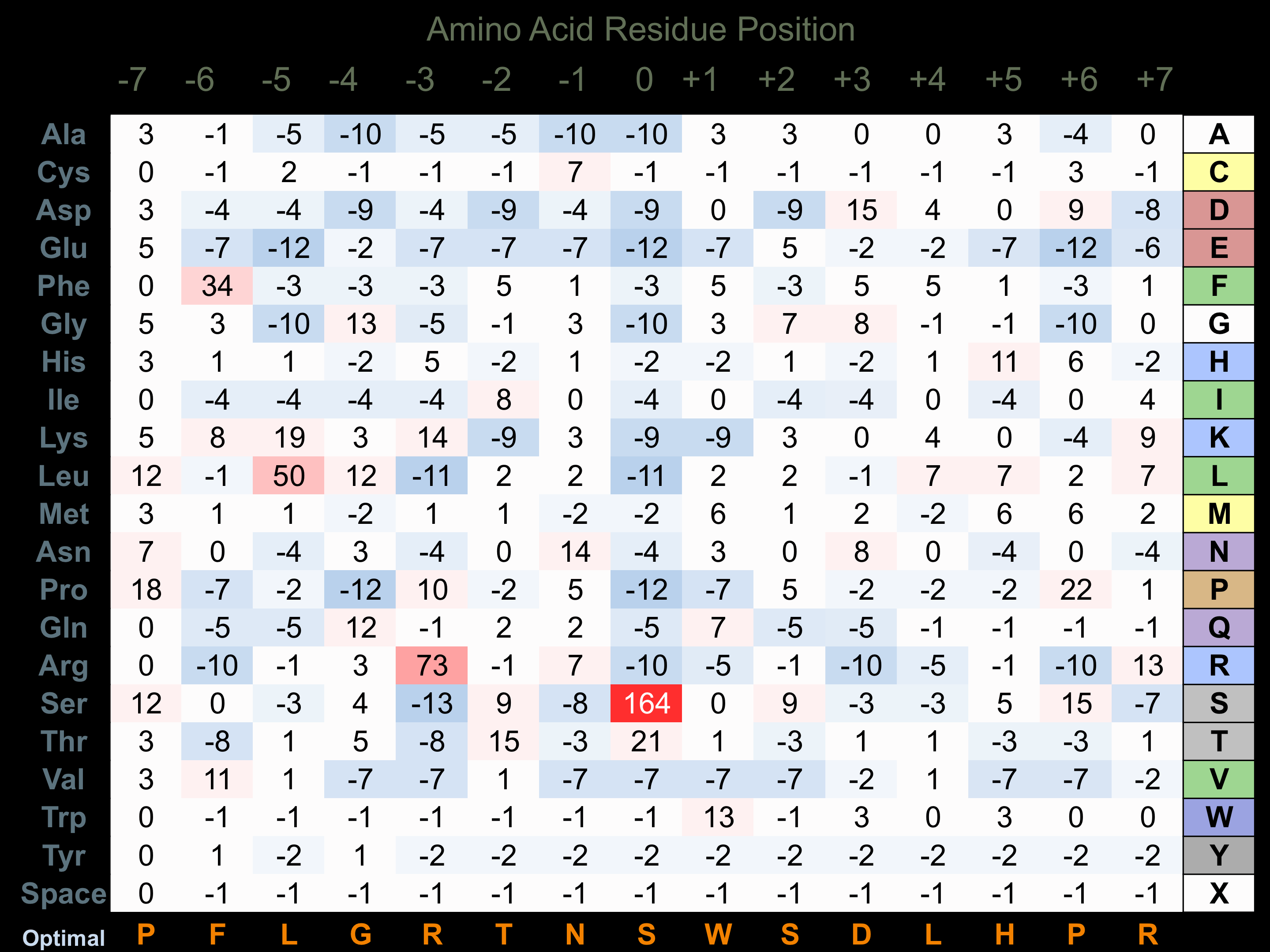
Matrix Type:
Experimentally derived from alignment of 33 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Oxygen inadequacy
Specific Diseases (Non-cancerous):
Hypoxia
Comments:
CHK1 appears to be a tumour suppressor protein (TSP). The active form of the protein kinase normally acts to inhibit tumour cell proliferation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -48, p<0.008); Breast sporadic basal-like cancer (BLC) (%CFC= +56, p<0.0001); Cervical cancer (%CFC= -47, p<0.0001); Cervical cancer stage 1B (%CFC= +578, p<0.018); Cervical cancer stage 2A (%CFC= +289, p<0.059); Classical Hodgkin lymphomas (%CFC= +51, p<0.0005); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +326, p<0.002); Colon mucosal cell adenomas (%CFC= +154, p<0.0001); Large B-cell lymphomas (%CFC= +78, p<(0.0003); Oral squamous cell carcinomas (OSCC) (%CFC= +77, p<0.0005); Ovary adenocarcinomas (%CFC= +315, p<0.0001); and Prostate cancer - primary (%CFC= +88, p<0.0001). The COSMIC website notes an up-regulated expression score for CHK1 in diverse human cancers of 485, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 59 for this protein kinase in human cancers was 1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. S345A mutation of CHK1 can lead to reduced phosphorylation and impaired activation to nuclear retention after checkpoint activation. Mutations at amino acid residues 372, 376, and 379 can lead to misfolding and binding with ubiquitin.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25545 diverse cancer specimens. This rate is only -16 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R160H (3); N63H (3).
Comments:
Six deletions (5 T226fs*14 deletionsframeshift mutations in kinase catalytic domain), 3 insertions (2 at T226fs*19 insertions frameshift) and no complex mutations are noted on the COSMIC website.