Nomenclature
Short Name:
CK1a1
Full Name:
Casein kinase I, alpha isoform
Alias:
- Casein kinase 1, alpha 1
- Casein kinase I, alpha isoform
- EC 2.7.11.1
- KC1A
- Kinase CK1-alpha
- CK1
- CK1-alpha
- CKI-alpha
- CSNK1A1
Classification
Type:
Protein-serine/threonine kinase
Group:
CK1
Family:
CK1
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
38,915
# Amino Acids:
337
# mRNA Isoforms:
3
mRNA Isoforms:
41,937 Da (365 AA; P48729-2); 38,915 Da (337 AA; P48729); 37,567 Da (325 AA; P48729-3)
4D Structure:
Monomer. Interacts with the Axin complex.
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 17 | 277 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
A2,K8.
Serine phosphorylated:
S3, S4, S105, S199, S218, S242, S311, S313, S332.
Threonine phosphorylated:
T184, T287, T321, T327.
Tyrosine phosphorylated:
Y17, Y32, Y59, Y85, Y292, Y294.
Ubiquitinated:
K62, K65, K138, K162, K225, K229, K302, K304, K325.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 92
92
1409
123
966
 17
17
264
41
214
 26
26
392
6
254
 35
35
527
401
603
 81
81
1237
123
755
 53
53
806
302
2118
 19
19
284
143
370
 100
100
1527
109
2589
 74
74
1134
45
629
 12
12
178
300
164
 13
13
206
57
184
 66
66
1005
471
717
 15
15
229
72
150
 16
16
247
24
218
 16
16
238
41
218
 15
15
230
73
244
 31
31
478
350
2311
 14
14
210
23
148
 19
19
297
291
262
 74
74
1125
479
781
 18
18
282
35
262
 17
17
266
47
229
 16
16
243
20
128
 20
20
305
23
199
 19
19
293
35
282
 70
70
1072
248
958
 21
21
317
75
192
 22
22
330
23
192
 21
21
319
23
200
 12
12
176
140
227
 45
45
680
60
425
 66
66
1010
126
1940
 22
22
335
213
510
 67
67
1022
317
767
 46
46
705
165
1183
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.8
98.8
98.8
100 97.6
97.6
98.2
100 -
-
-
100 -
-
-
100 99.7
99.7
99.7
100 -
-
-
- 57.2
57.2
67.8
100 96.1
96.1
96.1
100 -
-
-
- -
-
-
- 99.7
99.7
100
100 57.6
57.6
68
100 59.5
59.5
70.2
99 -
-
-
- 74.2
74.2
82.8
79 80.9
80.9
86.9
- 81.2
81.2
86.8
79 78.7
78.7
87.4
- -
-
-
- -
-
-
- -
-
-
72 50.2
50.2
63.1
67 41.9
41.9
54.7
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PER3 - P56645 |
| 2 | PER1 - O15534 |
| 3 | ARFGAP1 - Q8N6T3 |
| 4 | CTNNB1 - P35222 |
| 5 | TP53 - P04637 |
| 6 | BACE1 - P56817 |
| 7 | GJA1 - P17302 |
| 8 | CDK5 - Q00535 |
| 9 | PRKD2 - Q9BZL6 |
| 10 | SNCA - P37840 |
| 11 | DVL1 - O14640 |
| 12 | DVL1L1 - P54792 |
| 13 | PER2 - O15055 |
| 14 | PSEN2 - P49810 |
| 15 | PPP1R14A - Q96A00 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
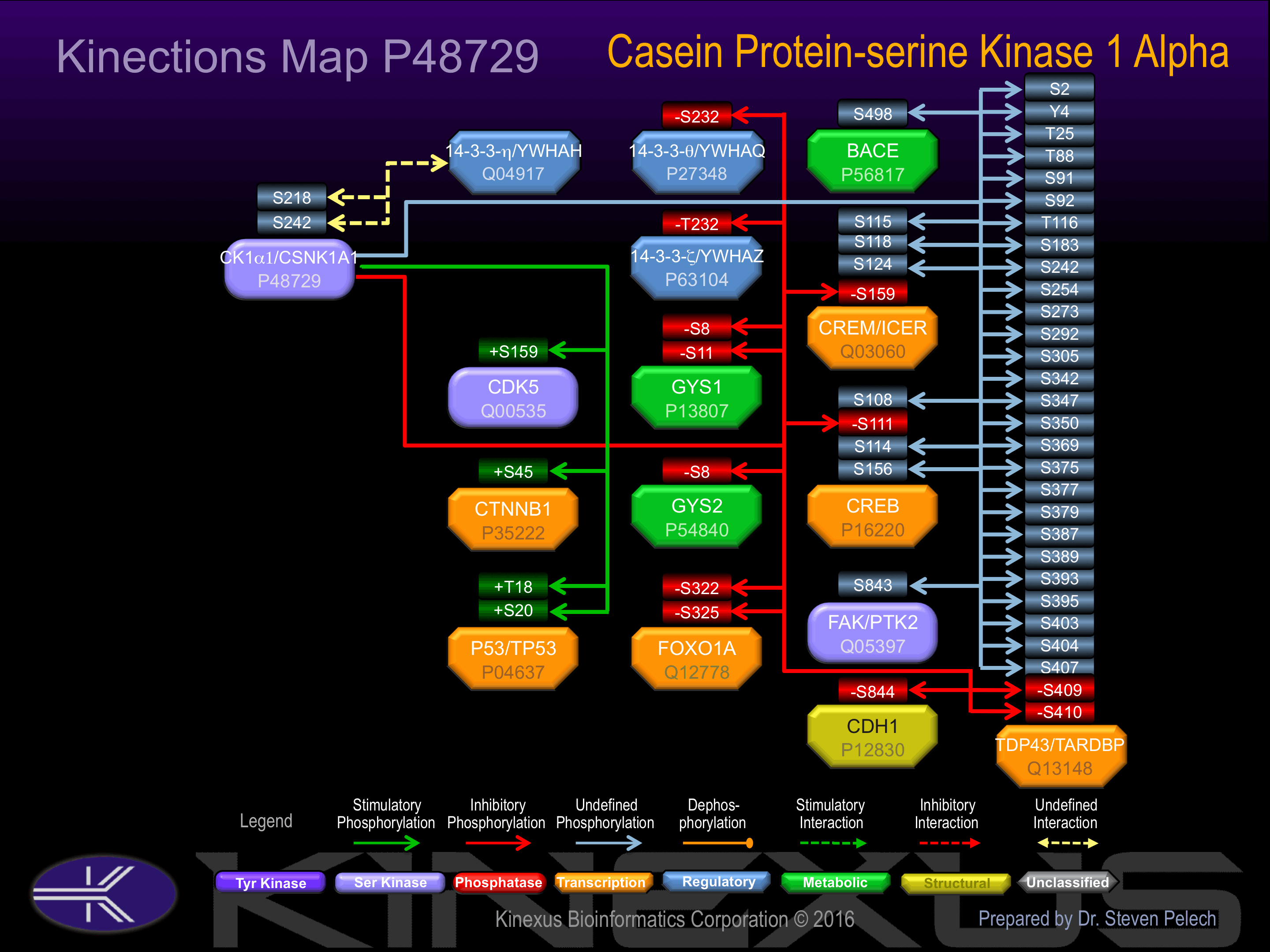
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 14-3-3 theta | P27348 | S232 | LTLWTSDSAGEECDA | - |
| 14-3-3 zeta (YWHAZ) | P63104 | T232 | LTLWTSDTQGDEAEA | - |
| AQP2 | P41181 | S256 | REVRRRQSVELHSPQ | |
| BACE | P56817 | S498 | DDFADDISLLK____ | |
| Bid | P55957 | T59 | EGYDELQTDGNRSSH | |
| Calsenilin | Q9Y2W7 | S63 | APQGSDSSDSELELS | |
| CDH1 | P12830 | S844 | GSGSEAASLSSLNSS | - |
| CDK5 | Q00535 | S159 | GIPVRCYSAEVVTLW | + |
| CREB1 | P16220 | S108 | QISTIAESEDSQESV | ? |
| CREB1 | P16220 | S111 | TIAESEDSQESVDSV | - |
| CREB1 | P16220 | S114 | ESEDSQESVDSVTDS | ? |
| CREB1 | P16220 | S156 | PRIEEEKSEEETSAP | |
| CREM | Q03060 | S115 | AIAETDESAESEGVI | |
| CREM | Q03060 | S118 | ETDESAESEGVIDSH | |
| CREM | Q03060 | S124 | ESEGVIDSHKRREIL | |
| CREM | Q03060 | S159 | PKIEEERSEEEGTPP | - |
| CTNNB1 | P35222 | S45 | GATTTAPSLSGKGNP | + |
| Cx43 | P17302 | S324 | NRMGQAGSTISNSHA | |
| Cx43 | P17302 | S327 | GQAGSTISNSHAQPF | |
| Cx43 | P17302 | S329 | AGSTISNSHAQPFDF | |
| DARPP-32 | Q9UD71 | S137 | EEEEEEDSQAEVLKV | |
| ECE1 | P42892 | S34 | DEEDLVDSLSEGDAY | |
| ECE1 | P42892 | S36 | EDLVDSLSEGDAYPN | |
| eIF2B-e | Q13144 | S469 | DGEFSDDSGADQEKD | |
| FADD | Q13158 | S194 | QNRSGAMSPMSWNSD | |
| FAK (PTK2) iso5 | Q05397 | S843 | DVRLSRGSIDREDGS | |
| FOXG1 | P55316 | S19 | MIPKSSFSINSLVPE | ? |
| FOXO1A | Q12778 | S322 | PRTSSNASTISGRLS | - |
| FOXO1A | Q12778 | S325 | SSNASTISGRLSPIM | - |
| Galectin-3 | P17931 | S11 | FSLHDALSGSGNPNP | |
| Galectin-3 | P17931 | S5 | ___ADNFSLHDALSG | |
| GRP94 | P14625 | S347 | KPIWQRPSKEVEEDE | |
| GYS1 | P13807 | S11 | NRTLSMSSLPGLEDW | - |
| GYS1 | P13807 | S8 | MPLNRTLSMSSLPGL | - |
| GYS2 | P54840 | S8 | MLRGRSLSVTSLGGL | - |
| HDAC1 | Q13547 | S421 | IACEEEFSDSEEEGE | - |
| hnRNP C1,C2 | P07910 | S253 | ETNVKMESEGGADDS | |
| hnRNP C1,C2 | P07910 | S260 | SEGGADDSAEEGDLL | |
| hnRNP C1,C2 | P07910 | S299 | EGEDDRDSANGEDDS | |
| IRS1 | P35568 | Y46 | GGPARLEYYENEKKW | |
| LRP6 | O75581 | T1493 | NPPPSPATERSHYTM | |
| MDM4 | O15151 | S289 | DDLEDSKSLSDDTDV | |
| NFAT4 | Q12968 | S177 | ISSRSWFSDASSCES | |
| NFAT4 | Q12968 | S180 | RSWFSDASSCESLSH | |
| NFAT4 | Q12968 | S181 | SWFSDASSCESLSHI | |
| NFAT4 | Q12968 | S184 | SDASSCESLSHIYDD | |
| NFAT4 | Q12968 | S186 | ASSCESLSHIYDDVD | |
| NFAT5 | O94916 | S176 | LLDNSRMSCQDEGCG | |
| p53 | P04637 | S20 | PLSQETFSDLWKLLP | + |
| p53 | P04637 | T18 | EPPLSQETFSDLWKL | + |
| Parkin | O60260 | S101 | GCEREPQSLTRVDLS | |
| Parkin | O60260 | S378 | AYHEGECSAVFEASG | |
| PRP1 | P02810 | S24 | QDLDEDVSQEDVPLV | |
| PRP1 | P02810 | S38 | VISDGGDSEQFIDEE | |
| RhoB | P62745 | S185 | ALQKRYGSQNGCINC | |
| SLC4A1 | P02730 | S303 | IDAYMAQSRGELLHS | |
| SLC4A1 | P02730 | T42 | AAHDTEATATDYHTT | |
| SNCA | P37840 | S129 | NEAYEMPSEEGYQDY | |
| TARDBP | Q13148 | S183 | KLPNSKQSQDEPLRS | |
| TARDBP | Q13148 | S2 | ______MSEYIRVTE | |
| TARDBP | Q13148 | S242 | ADDQIAQSLCGEDLI | |
| TARDBP | Q13148 | S254 | DLIIKGISVHISNAE | |
| TARDBP | Q13148 | S273 | SNRQLERSGRFGGNP | |
| TARDBP | Q13148 | S292 | NQGGFGNSRGGGAGL | |
| TARDBP | Q13148 | S305 | GLGNNQGSNMGGGMN | |
| TARDBP | Q13148 | S342 | GMMGMLASQQNQSGP | |
| TARDBP | Q13148 | S347 | LASQQNQSGPSGNNQ | |
| TARDBP | Q13148 | S350 | QQNQSGPSGNNQNQG | |
| TARDBP | Q13148 | S369 | EPNQAFGSGNNSYSG | |
| TARDBP | Q13148 | S375 | GSGNNSYSGSNSGAA | |
| TARDBP | Q13148 | S377 | GNNSYSGSNSGAAIG | |
| TARDBP | Q13148 | S379 | NSYSGSNSGAAIGWG | |
| TARDBP | Q13148 | S387 | GAAIGWGSASNAGSG | |
| TARDBP | Q13148 | S389 | AIGWGSASNAGSGSG | |
| TARDBP | Q13148 | S393 | GSASNAGSGSGFNGG | |
| TARDBP | Q13148 | S395 | ASNAGSGSGFNGGFG | |
| TARDBP | Q13148 | S403 | GFNGGFGSSMDSKSS | |
| TARDBP | Q13148 | S404 | FNGGFGSSMDSKSSG | |
| TARDBP | Q13148 | S407 | GFGSSMDSKSSGWGM | |
| TARDBP | Q13148 | S409 | GSSMDSKSSGWGM__ | - |
| TARDBP | Q13148 | S410 | SSMDSKSSGWGM___ | - |
| TARDBP | Q13148 | S91 | KMDETDASSAVKVKR | |
| TARDBP | Q13148 | S92 | MDETDASSAVKVKRA | |
| TARDBP | Q13148 | T116 | LGLPWKTTEQDLKEY | |
| TARDBP | Q13148 | T25 | IPSEDDGTVLLSTVT | |
| TARDBP | Q13148 | T88 | NKRKMDETDASSAVK | |
| TARDBP | Q13148 | Y4 | ____MSEYIRVTEDE | |
| TBX21 | Q9UL17 | S513 | PSSGDSSSPAGAPSP | |
| VHL | P40337 | S72 | SVNSREPSQVIFCNR | |
| VMAT2 | Q05940 | S511 | PIGEDEESESD____ | |
| VMAT2 | Q05940 | S513 | GEDEESESD______ |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
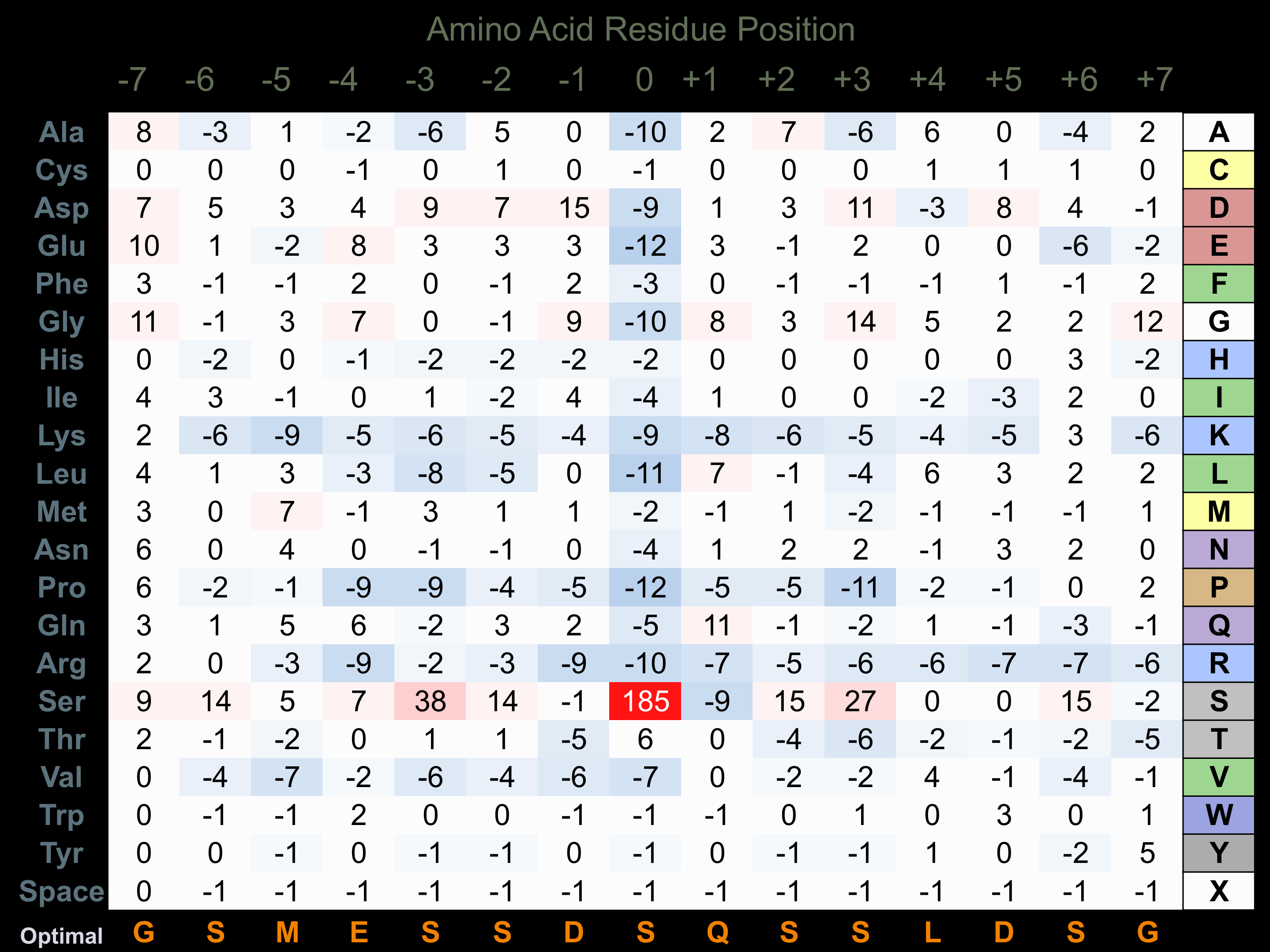
Matrix Type:
Experimentally derived from alignment of 122 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Lung cancer (LC)
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +138, p<0.0001); Brain glioblastomas (%CFC= -62, p<0.009); Brain oligodendrogliomas (%CFC= -77, p<0.002); Cervical cancer (%CFC= +45, p<0.003); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -71, p<0.0001); Skin squamous cell carcinomas (%CFC= +65, p<0.038); and Vulvar intraepithelial neoplasia (%CFC= +70, p<0.002). The COSMIC website notes an up-regulated expression score for CK1a1 in diverse human cancers of 407, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 314 for this protein kinase in human cancers was 5.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25239 diverse cancer specimens. This rate is only -4 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: D140A (4), A185V (4). These are located in the kinase catalytic domain.
Comments:
Only 2 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.