Nomenclature
Short Name:
CaMK1d
Full Name:
Calcium/calmodulin-dependent protein kinase type I delta
Alias:
- CAMK1D
- CaMK1-delta
- CaM-KI delta
- EC 2.7.11.17
- KCC1D
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMK1
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
42,914
# Amino Acids:
385
# mRNA Isoforms:
2
mRNA Isoforms:
42,914 Da (385 AA; Q8IU85); 40,190 Da (357 AA; Q8IU85-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
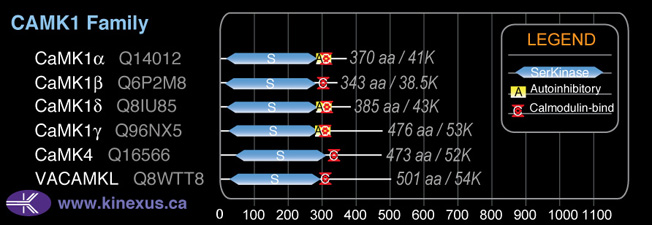
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 23 | 279 | Pkinase |
| 279 | 319 | Autoinhibitory |
| 299 | 320 | CaM_binding |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K174, K196, K262.
Serine phosphorylated:
S8, S122+, S179, S326, S327, S338.
Threonine phosphorylated:
T180+, T184.
Tyrosine phosphorylated:
Y152, Y153.
Ubiquitinated:
K52.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 56
56
1129
22
1130
 4
4
71
9
37
 5
5
98
8
65
 19
19
385
82
435
 27
27
540
24
502
 3
3
58
46
62
 3
3
51
25
41
 16
16
320
24
493
 10
10
205
10
175
 3
3
59
64
59
 3
3
63
18
45
 36
36
730
76
594
 3
3
69
19
84
 2
2
32
6
12
 6
6
129
15
128
 1.2
1.2
25
12
14
 45
45
910
88
5338
 3
3
68
13
49
 2
2
38
62
23
 37
37
750
84
687
 4
4
89
15
69
 4
4
87
17
73
 3
3
59
8
39
 4
4
72
13
55
 9
9
187
15
130
 38
38
754
50
964
 4
4
75
22
88
 4
4
88
13
58
 3
3
53
12
28
 5
5
102
28
93
 51
51
1030
30
520
 100
100
2008
26
3536
 8
8
161
69
364
 42
42
845
52
716
 5
5
99
53
139
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
99.7
100 56.8
56.8
59.8
96 -
-
-
98 -
-
-
- 73.3
73.3
78.2
98 -
-
-
- 97.7
97.7
99.2
98 72.2
72.2
80.3
98 -
-
-
- 86.3
86.3
88.7
- 57.6
57.6
68.3
93 41.6
41.6
61.8
87 38.1
38.1
58.5
89 37.3
37.3
57.5
- 29.4
29.4
45.9
60 -
-
-
- -
-
-
66 -
-
-
- -
-
-
- 31.2
31.2
46.2
- -
-
-
42 29.1
29.1
43.5
40 -
-
-
42 -
-
-
41
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
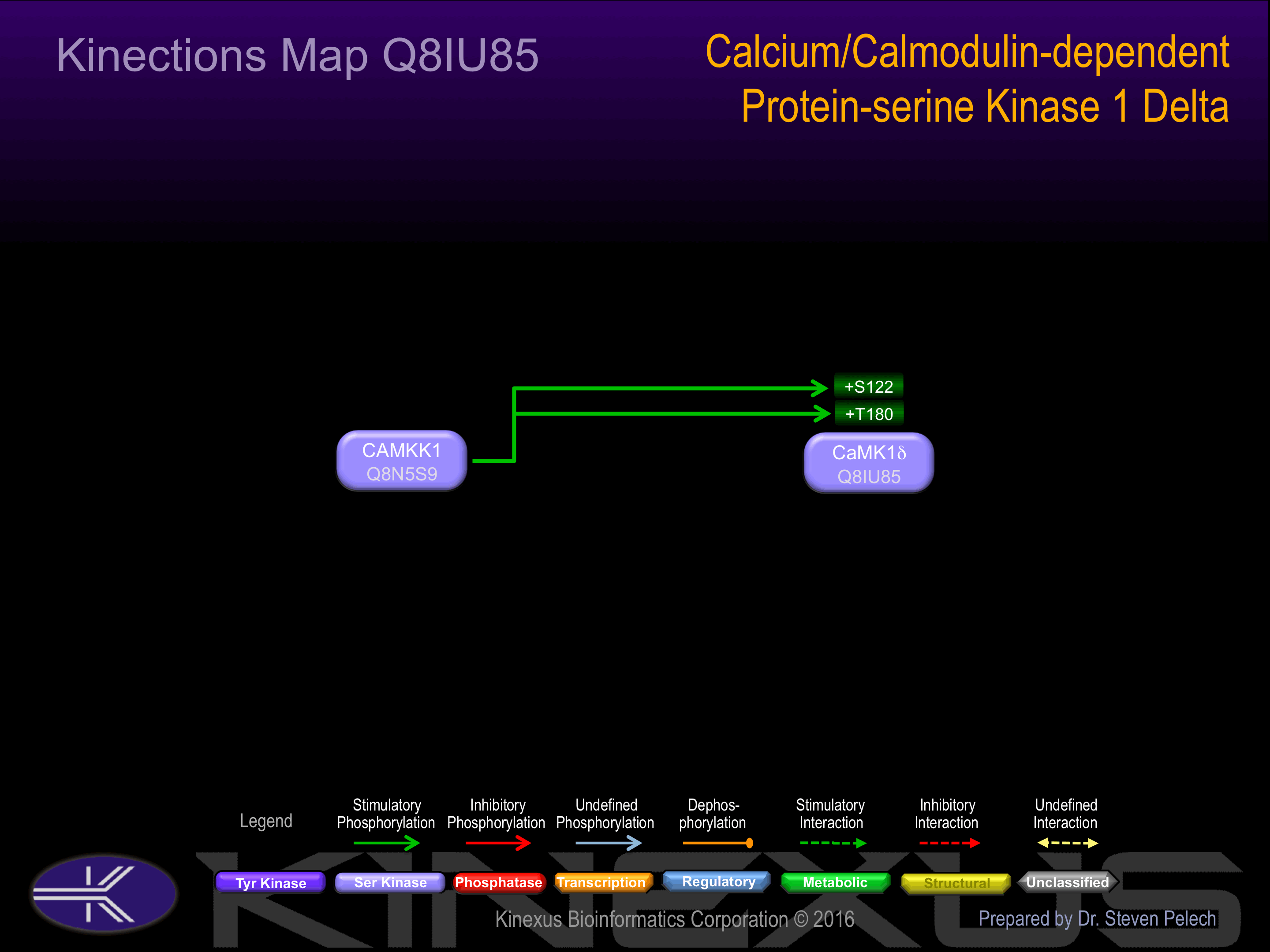
Activated by Ca2+/calmodulin. Binding of calmodulin is thought to result in a conformational change and can lead to subsequent activation through phosphorylation by CAMKK1 and CAMKK2. At least in part, can also be activated by CAMKK1 in a calcium-independent manner.
Inhibition:
NA
Synthesis:
Expression increases upon treatment of EC cells with DMSO and retinoic acid.
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
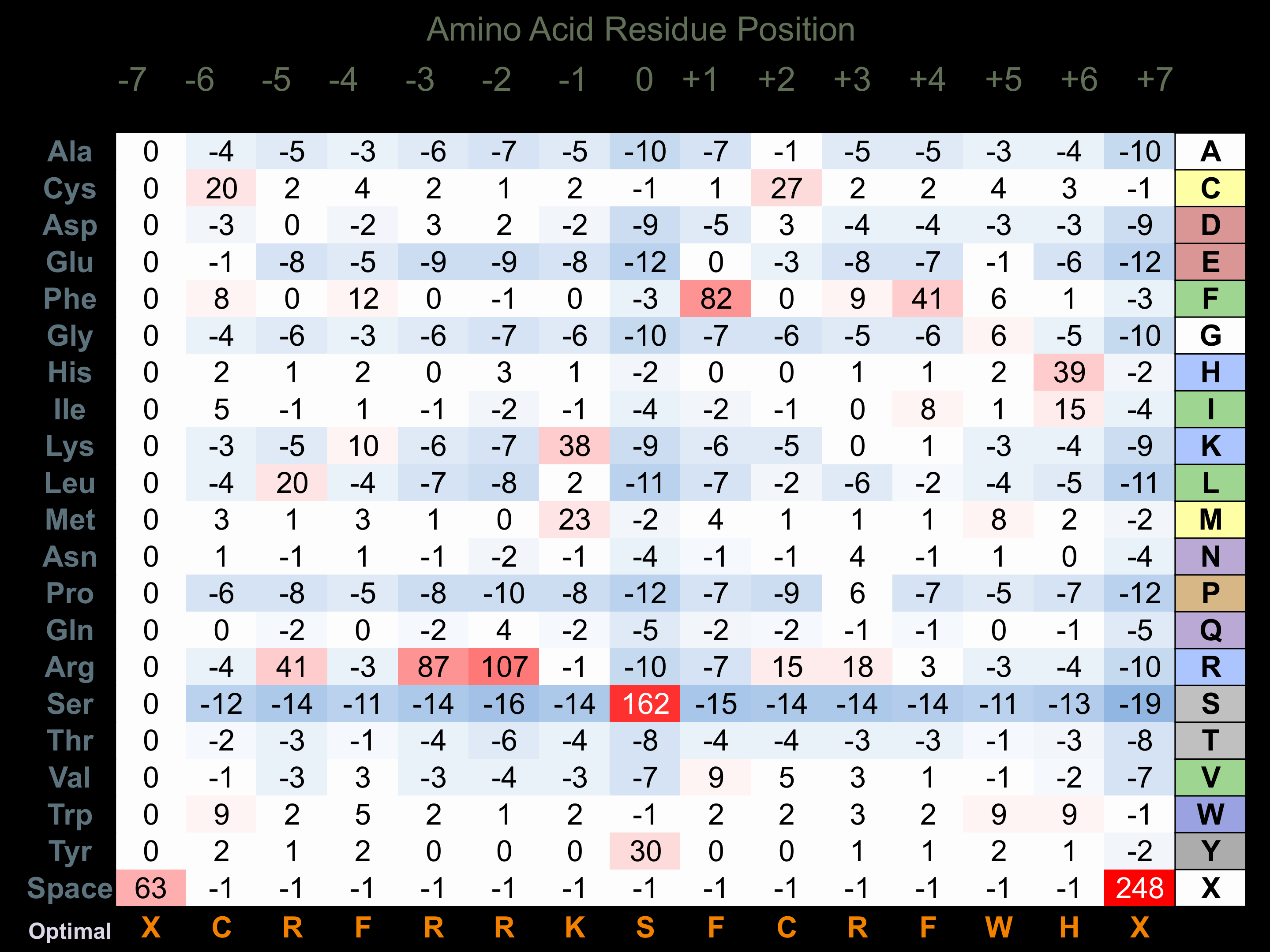
Matrix Type:
Derived from alignment of 32 peptides phosphorylated by recombinant CaMK1-delta in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
Comments:
A T180A mutation in CaMK1d leads to loss of ionomycin-induced activation. CaMK1d may play a role in the apoptosis of erythroleukemia cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -71, p<0.01); Cervical cancer (%CFC= +47, p<0.074); Clear cell renal cell carcinomas (cRCC) (%CFC= +81, p<0.022); and Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +668, p<0.003). The COSMIC website notes an up-regulated expression score for CaMK1d in diverse human cancers of 442, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 24757 diverse cancer specimens. This rate is only 30 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.73 % in 895 skin cancers tested; 0.49 % in 1270 large intestine cancers tested; 0.39 % in 603 endometrium cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,009 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.