Nomenclature
Short Name:
CaMK2d
Full Name:
Calcium-calmodulin-dependent protein kinase type II delta chain
Alias:
- Calcium/calmodulin-dependent protein kinase II delta
- Calcium/calmodulin-dependent protein kinase type II delta chain
- CaM-kinase II delta
- EC 2.7.11.17
- KCC2D
- KCCD
- CaM kinase II delta
- CaMK2-delta
- CAMKD
- CaMK-II delta
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMK2
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
56369
# Amino Acids:
499
# mRNA Isoforms:
10
mRNA Isoforms:
59,152 Da (524 AA; Q13557-11); 58,472 Da (519 AA; Q13557-4); 57,901 Da (513 AA; Q13557-6); 57,620 Da (510 AA; Q13557-3); 56,369 Da (499 AA; Q13557); 56,230 Da (498 AA; Q13557-5); 55,660 Da (492 AA; Q13557-10); 55,378 Da (489 AA; Q13557-9); 54,128 Da (478 AA; Q13557-8); 54,114 Da (478 AA; Q13557-12)
4D Structure:
CAMK2 is composed of four different chains: alpha, beta, gamma, and delta. The different isoforms assemble into homo- or heteromultimeric holoenzymes composed of 8 to 12 subunits. Interacts with RRAD
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
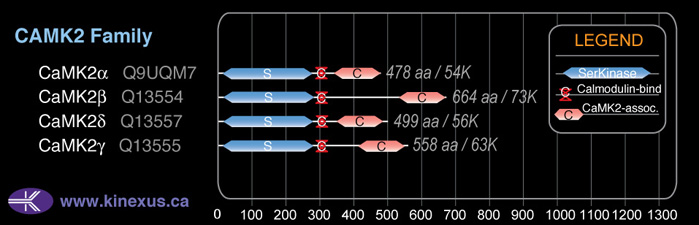
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 14 | 272 | Pkinase |
| 297 | 312 | CaM_binding |
| 346 | 473 | CaMK2-Association |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
A2, K227.
Methylated:
K301.
Serine phosphorylated:
S3, S79, S146, S235, S276, S280, S315, S319, S330, S333, S334, S404, S441, S472, S490, S494.
Threonine phosphorylated:
T254, T287+, T306, T307, T311, T331, T336, T337.
Tyrosine phosphorylated:
Y231.
Ubiquitinated:
K43, K227, K251.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
2050
33
1482
 4
4
84
8
82
 26
26
526
15
396
 30
30
617
157
830
 45
45
917
51
634
 23
23
476
53
1039
 10
10
199
53
206
 50
50
1026
36
1505
 3
3
64
3
2
 6
6
122
129
97
 8
8
158
24
142
 30
30
614
85
662
 15
15
317
15
226
 3
3
61
3
3
 7
7
135
24
161
 4
4
76
25
61
 12
12
242
226
1531
 5
5
111
18
142
 23
23
469
108
494
 47
47
972
143
631
 14
14
294
24
300
 10
10
207
24
207
 9
9
186
31
254
 12
12
239
18
236
 6
6
133
24
169
 44
44
912
111
1422
 16
16
323
18
274
 10
10
203
18
167
 13
13
268
18
244
 5
5
93
70
83
 30
30
618
24
153
 21
21
426
31
1088
 19
19
384
92
836
 41
41
839
140
740
 6
6
130
92
283
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 94.6
94.6
95.2
100 93
93
93.4
99 -
-
-
100 -
-
-
- 90.8
90.8
91.2
98 -
-
-
- 100
100
100
100 92.7
92.7
93.3
99 -
-
-
- -
-
-
- 94.6
94.6
95.6
99 92.2
92.2
94.2
97 87.4
87.4
92
92 -
-
-
- 74.2
74.2
82.5
81 -
-
-
- -
-
-
73 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 30.2
30.2
47.7
39 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | RPS18 - P62269 |
| 2 | VIM - P08670 |
| 3 | EIF4B - P23588 |
| 4 | CAMK2B - Q13554 |
| 5 | GRIN2B - Q13224 |
| 6 | CAMK2A - Q9UQM7 |
| 7 | LRRC7 - Q96NW7 |
Regulation
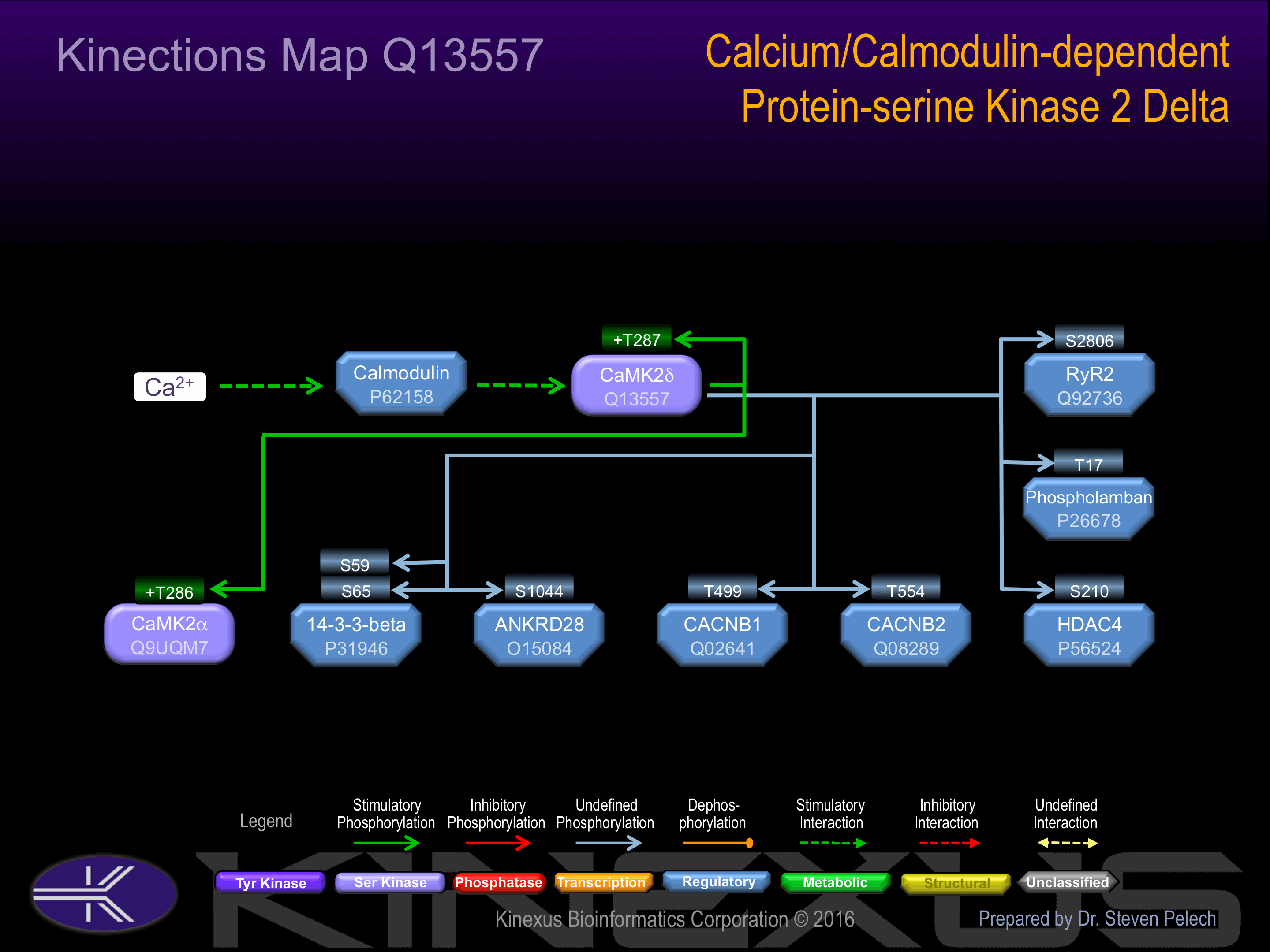
Activation:
Activated in response to a rise in intracellular Ca(2+) concentration. Autophosphorylation at Thr-287 probably increases phosphotransferase activity in a Ca(2+)-independent manner.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 14-3-3 beta (YWHAB) | P31946 | S59 | NVVGARRSSWRVISS | |
| 14-3-3 beta (YWHAB) | P31946 | S65 | SSWRVISSIEQKTER | |
| ANKRD28 | O15084 | S1044 | TNTSKTVSFEALPIM | |
| CACNB1 | Q02641 | T499 | RALSRQDTFDADTPG | |
| CACNB2 | Q08289 | T554 | RGLSRQETFDSETQE | |
| CaMK2a | Q9UQM7 | T286 | SCMHRQETVDCLKKF | + |
| CaMK2d | Q13557 | T287 | SMMHRQETVDCLKKF | + |
| HDAC4 | P56524 | S210 | YGKTQHSSLDQSSPP | |
| Phospholamban | P26678 | T17 | SAIRRASTIEMPQQA | |
| RyR2 | Q92736 | S2806 | YNRTRRISQTSQVSV |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
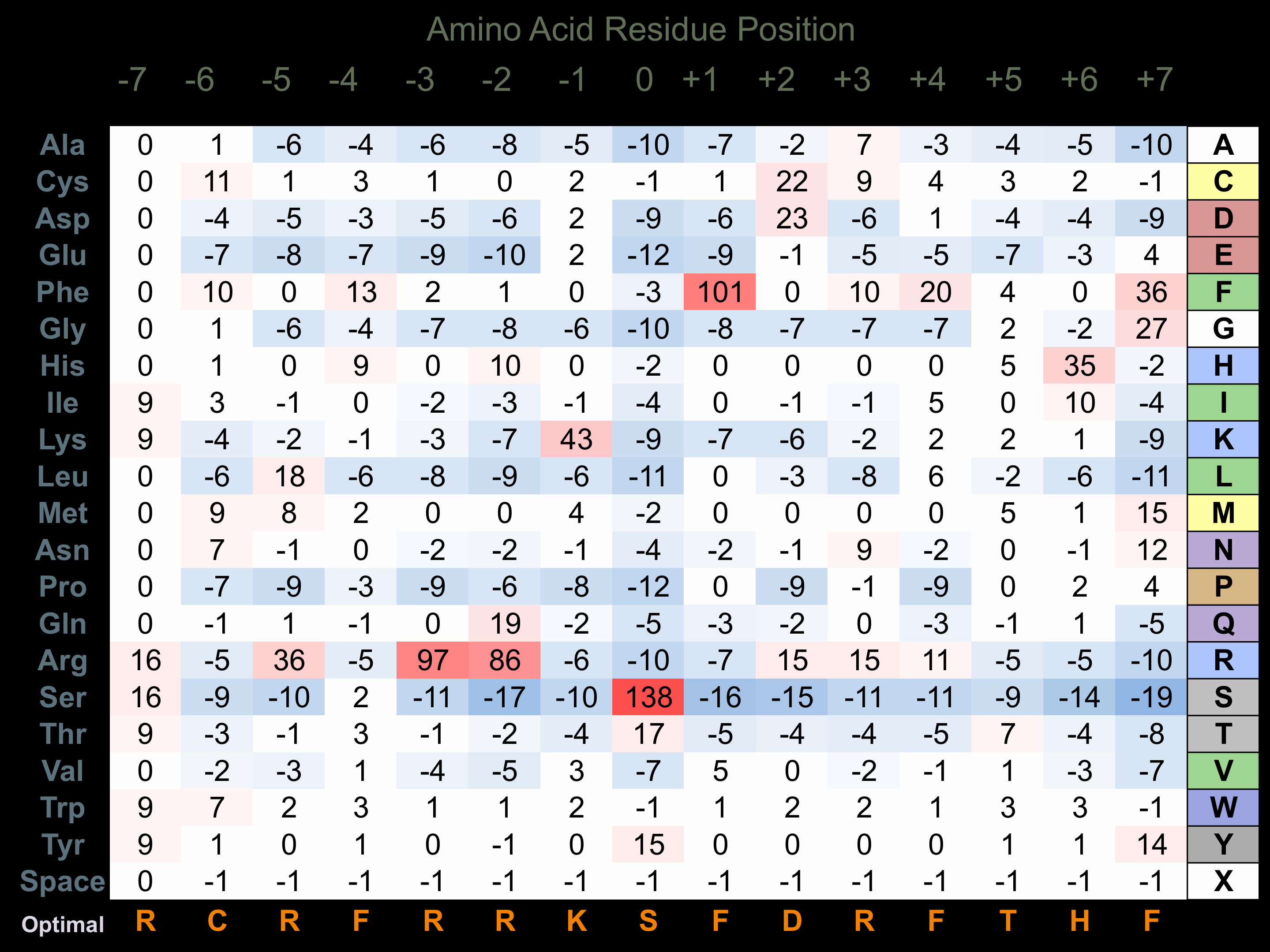
Matrix Type:
Experimentally derived from alignment of 9 known protein substrate phosphosites and 31 peptides phosphorylated by recombinant CaMK2-delta in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Heart disorders
Specific Diseases (Non-cancerous):
Dilated cardiomyopathy
Comments:
From semiquantitative RT-PCR of left ventricular tissues from normal hearts and from patients suffering from dilated cardiomyopathy, expression of the major cardiac CAMK2d transcript was significantly increased with disease.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -50, p<0.075); Cervical cancer stage 2A (%CFC= -48, p<0.019); Ovary adenocarcinomas (%CFC= -58, p<0.007); Prostate cancer (%CFC= -59, p<0.019); Prostate cancer - metastatic (%CFC= +48, p<0.074). The COSMIC website notes an up-regulated expression score for CAMK2d in diverse human cancers of 301, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 32 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24726 diverse cancer specimens. This rate is only -20 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.26 % in 1093 large intestine cancers tested; 0.25 % in 805 skin cancers tested; 0.14 % in 1619 lung cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,034 cancer specimens
Comments:
Only 2 insertions and no insertions or complex mutations are noted on the COSMIC website.