Nomenclature
Short Name:
CaMK2g
Full Name:
Calcium-calmodulin-dependent protein kinase type II gamma chain
Alias:
- Calcium/calmodulin-dependent protein kinase II gamma
- CaM kinase II gamma subunit
- CaM-kinase II gamma
- CaM-kinase II gamma chain
- EC 2.7.11.17
- Kinase CaMK2-gamma
- CaMK2-gamma
- CAMKG
- CaMK-II gamma
- CaMK-II gamma subunit
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMK2
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
62,609
# Amino Acids:
558
# mRNA Isoforms:
10
mRNA Isoforms:
65,242 Da (588 AA; Q13555-6); 62,609 Da (558 AA; Q13555); 62,165 Da (556 AA; Q13555-8); 61,368 Da (547 AA; Q13555-2); 60,743 Da (542 AA; Q13555-9); 59,760 Da (533 AA; Q13555-7); 59,607 Da (529 AA; Q13555-4); 59,038 Da (527 AA; Q13555-5); 58,963 Da (524 AA; Q13555-3); 55,961 Da (495 AA; Q13555-10)
4D Structure:
CAMK2 is composed of four different chains: alpha, beta, gamma, and delta. The different isoforms assemble into homo- or heteromultimeric holoenzymes composed of 8 to 12 subunits.
1D Structure:
Subfamily Alignment
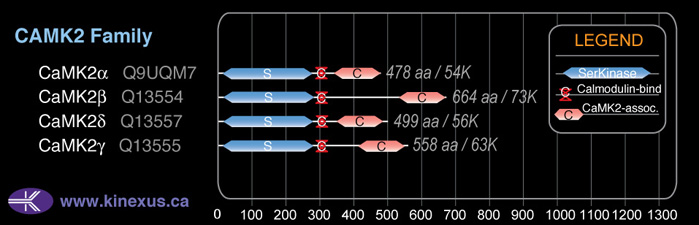
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 14 | 272 | Pkinase |
| 291 | 301 | CaM_binding |
| 426 | 553 | CaMK2-Association |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K227.
Serine phosphorylated:
S26-, S79, S114, S146, S235, S276, S280, S311, S315, S319, S325, S334, S349, S352, S384, S419, S484.
Threonine phosphorylated:
T35, T254, T287+, T306, T382, T388.
Tyrosine phosphorylated:
Y14, Y231.
Ubiquitinated:
K227, K246.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 55
55
907
42
1011
 5
5
89
23
57
 18
18
297
3
179
 53
53
877
128
580
 40
40
655
36
569
 4
4
72
111
34
 31
31
521
55
632
 51
51
848
49
1224
 34
34
565
24
382
 12
12
192
141
134
 7
7
117
36
107
 38
38
635
298
656
 6
6
99
36
46
 5
5
89
20
49
 10
10
165
28
237
 5
5
76
23
48
 12
12
196
376
634
 14
14
234
16
175
 23
23
383
168
280
 31
31
507
162
524
 25
25
417
28
347
 10
10
168
28
141
 4
4
65
22
65
 5
5
88
16
58
 7
7
122
26
124
 47
47
774
70
948
 5
5
87
39
44
 8
8
130
17
59
 14
14
227
20
120
 17
17
287
42
149
 48
48
790
30
793
 100
100
1654
51
3336
 8
8
139
88
226
 43
43
715
88
685
 6
6
102
48
48
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 86.1
86.1
87.8
100 -
-
-
100 -
-
-
96 94.7
94.7
94.7
100 -
-
-
- 94.6
94.6
94.8
100 86.3
86.3
87.9
98 -
-
-
- 84.4
84.4
85.4
- 76.5
76.5
81.7
94 75.2
75.2
80.4
96 75.2
75.2
82.4
88 -
-
-
- 69.7
69.7
81.3
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
57 28.4
28.4
47.1
32 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | RRAD - P55042 |
| 2 | CHAT - P28329 |
| 3 | GRIA1 - P42261 |
| 4 | SPR - P35270 |
| 5 | FLNA - P21333 |
| 6 | EGFR - P00533 |
| 7 | SYN1 - P17600 |
| 8 | CALM1 - P62158 |
| 9 | TH - P07101 |
| 10 | PEA15 - Q15121 |
| 11 | MYLK - Q15746 |
| 12 | PLCB3 - Q01970 |
| 13 | LRRC7 - Q96NW7 |
| 14 | ACTN2 - P35609 |
| 15 | GRIN2B - Q13224 |
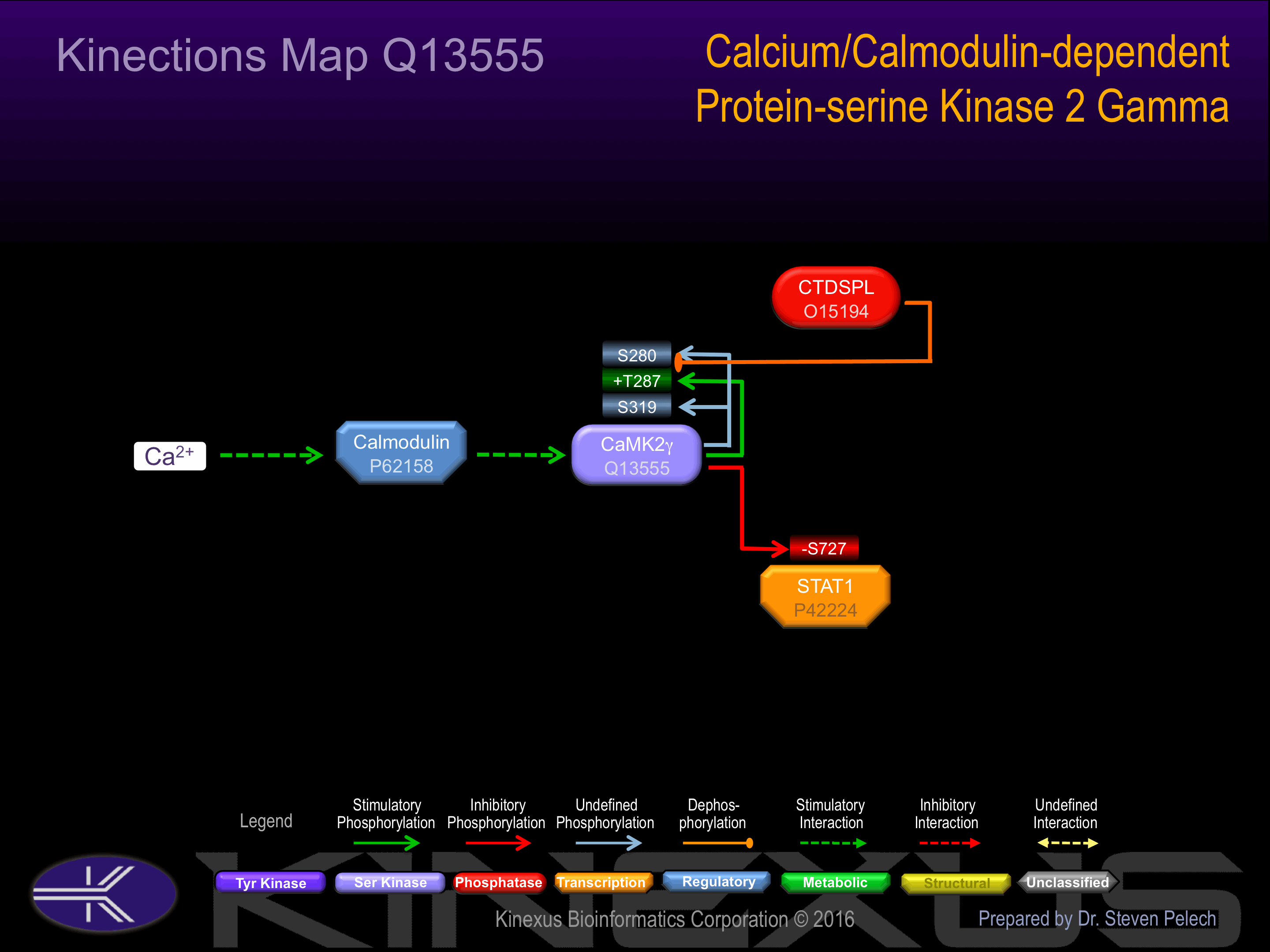
Regulation
Activation:
Activated in response to a rise in intracellular Ca(2+) concentration. Autophosphorylation at Thr-287 probably increases phosphotransferase activity in a Ca(2+)-independent manner.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
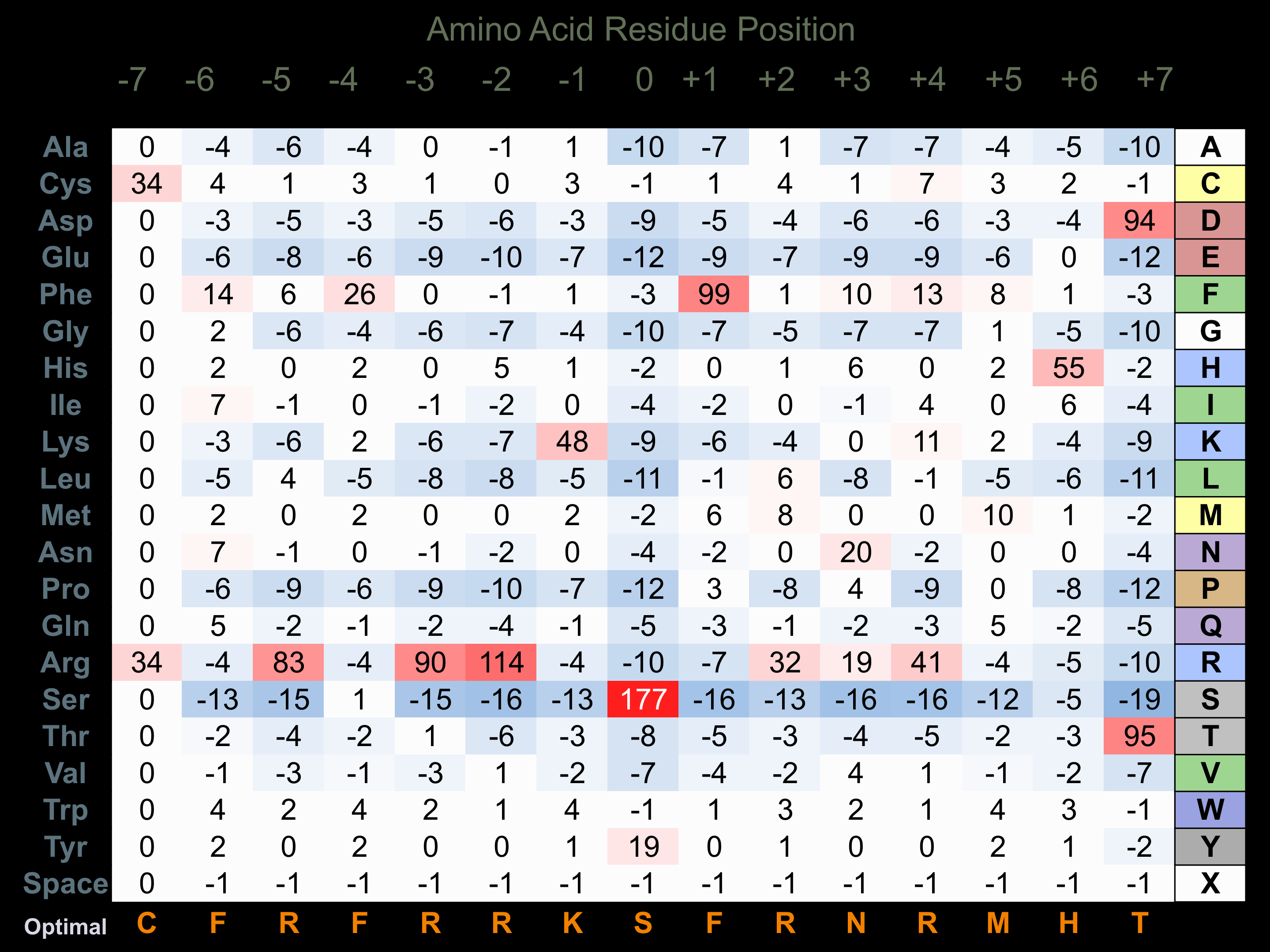
Matrix Type:
Experimentally derived from alignment of 2 known protein substrate phosphosites and 20 peptides phosphorylated by recombinant CAMK2-gamma in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, ischemia
Specific Diseases (Non-cancerous):
Ischemia; Timothy syndrome; Transient cerebral ischemia; Brain ischemia; Usher syndrome, Type 1f
Comments:
Usher syndrome type 1f is characterized by the loss of hearing and vision.
Specific Cancer Types:
Colorectal cancer (CRC)
Comments:
Timothy syndrome is related to colorectal cancer and long qt syndrome but will also lead to the malformation of cardiac, hand/foot, facial, and neuronal tissues.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +56, p<0.023); Colorectal adenocarcinomas (early onset) (%CFC= +48, p<0.09); Gastric cancer (%CFC= +149, p<0.0001); Ovary adenocarcinomas (%CFC= +55, p<0.073); and Skin melanomas (%CFC= -45, p<0.073). The COSMIC website notes an up-regulated expression score for CaMK2g in diverse human cancers of 476, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 114 for this protein kinase in human cancers was 1.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24751 diverse cancer specimens. This rate is only -38 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.25 % in 864 skin cancers tested; 0.24 % in 589 stomach cancers tested; 0.2 % in 548 urinary tract cancers tested; 0.2 % in 1270 large intestine cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,034 cancer specimens
Comments:
Only 1 insertion at C-terminus and no deletions or complex mutations are noted on the COSMIC website.