Nomenclature
Short Name:
CaMK4
Full Name:
Calcium-calmodulin-dependent protein kinase type IV
Alias:
- Brain Ca++-calmodulin-dependent protein kinase type IV
- Calcium/calmodulin-dependent protein kinase IV
- CAM kinase-GR
- CaMK IV
- CAMK4
- Kinase CaMK4; CaMK-GR; CaMKIV; EC 2.7.11.17; KCC4;
- Calcium/calmodulin-dependent protein kinase type IV catalytic chain
- Calspermin
- CAM kinase- GR
- CAM kinase IV
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
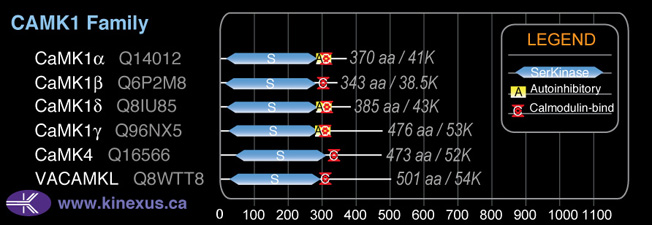
Family:
CAMK1
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
51,926
# Amino Acids:
473
# mRNA Isoforms:
1
mRNA Isoforms:
51,926 Da (473 AA; Q16566)
4D Structure:
Monomer. Interacts with serine/threonine protein phosphatase 2A catalytic subunit, PPP2CA or PPP2CB. The interaction with PP2CA or PP2CB is mutually exclusive with binding to Ca2+/calmodulin.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 46 | 300 | Pkinase |
| 323 | 342 | CaM_binding |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K190 (N6).
O-GlcNAcylated:
T57, S58, S137, S189, S344, S345, S356.
Serine phosphorylated:
S8, S12+, S13+, S15, S50, S137, S189-, S336-, S337, S341, S343, S344, S345, S352, S356, S360.
Threonine phosphorylated:
T200+, T204-.
Tyrosine phosphorylated:
Y32.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 56
56
1040
22
1281
 2
2
31
13
37
 2
2
41
11
38
 9
9
166
94
299
 18
18
337
23
268
 7
7
132
54
372
 16
16
297
37
423
 44
44
831
42
2597
 11
11
211
10
208
 2
2
45
99
50
 1.5
1.5
28
35
31
 15
15
284
171
451
 5
5
97
22
93
 1.5
1.5
28
13
27
 2
2
35
30
49
 0.9
0.9
16
15
17
 0.8
0.8
15
361
18
 1.4
1.4
26
19
29
 1.3
1.3
24
83
22
 16
16
300
84
275
 1.4
1.4
27
30
33
 2
2
32
32
32
 2
2
30
20
27
 7
7
132
22
81
 10
10
178
32
191
 100
100
1868
58
4710
 3
3
53
25
49
 2
2
31
22
36
 1.2
1.2
23
21
24
 7
7
135
28
148
 47
47
878
18
591
 79
79
1473
21
4872
 20
20
382
60
857
 28
28
514
57
486
 29
29
535
35
543
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 78.3
78.3
78.4
100 73.3
73.3
74.9
94.5 -
-
-
83 -
-
-
92 34
34
49.7
90 -
-
-
- 80.3
80.3
85.6
82 80
80
86.5
83 -
-
-
- 70.2
70.2
78
- 34.5
34.5
53.9
80 33.7
33.7
52.2
81 65.3
65.3
72.3
79 -
-
-
- 30.4
30.4
49.8
- -
-
-
- -
-
-
- 40.4
40.4
56.2
- -
-
-
- -
-
-
- -
-
-
43 32.2
32.2
48.6
42 -
-
-
- -
-
-
41
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CAMKK1 - Q8N5S9 |
| 2 | RELA - Q04206 |
| 3 | HDAC4 - P56524 |
| 4 | CREBBP - Q92793 |
| 5 | CABIN1 - Q9Y6J0 |
| 6 | NOS1 - P29475 |
| 7 | CAMKK2 - Q96RR4 |
| 8 | GAPDH - P04406 |
Regulation
Activation:
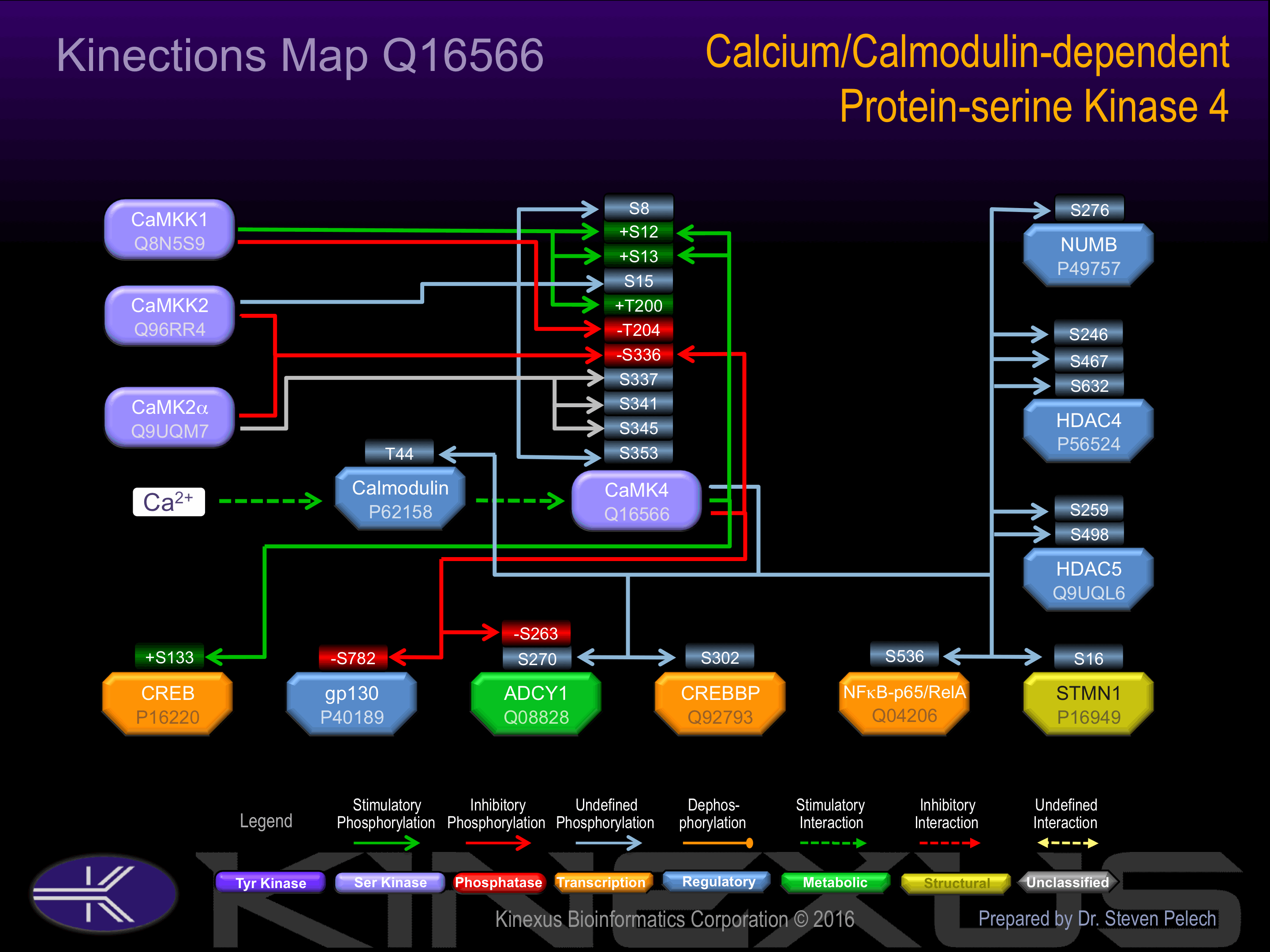
Activated by Ca2+/calmodulin. ; Must be phosphorylated to be maximally active. Phosphorylation at Ser-12, Ser-13 and Thr-200 increases phosphotransferase activity. Phosphorylated by CAMKK1 or CAMKK2. Autophosphorylation of the N-terminus is required for full activation. In part, activity is independent on Ca2+/calmodulin and autophosphorylation of Ser-336 allows to switch to a Ca2+/calmodulin-independent state
Inhibition:
Probably inactivated by serine/threonine protein phosphatase 2A.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| CaMKK2 | Q96RR4 | S8 | MLKVTVPSCSASSCS | |
| CaMKK1 | Q8N5S9 | S12 | TVPSCSASSCSSVTA | + |
| CaMK4 | Q16566 | S12 | TVPSCSASSCSSVTA | + |
| CaMKK1 | Q8N5S9 | S13 | VPSCSASSCSSVTAS | + |
| CaMK4 | Q16566 | S13 | VPSCSASSCSSVTAS | + |
| CaMKK2 | Q96RR4 | S15 | SCSASSCSSVTASAA | |
| CaMKK1 | Q8N5S9 | T200 | EHQVLMKTVCGTPGY | + |
| CaMKK1 | Q8N5S9 | T204 | LMKTVCGTPGYCAPE | - |
| CaMK2a | Q9UQM7 | S336 | AVKAVVASSRLGSAS | - |
| CaMK4 | Q16566 | S336 | AVKAVVASSRLGSAS | - |
| CaMK2a | Q9UQM7 | S337 | VKAVVASSRLGSASS | |
| CaMK2a | Q9UQM7 | S341 | VASSRLGSASSSHGS | |
| CaMK2a | Q9UQM7 | S345 | RLGSASSSHGSIQES |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ADCY1 | Q08828 | S263 | RRALRTASEKLRNRS | - |
| ADCY1 | Q08828 | S270 | SEKLRNRSSFSTNVV | |
| Calmodulin | P62158 | T44 | RSLGQNPTEAELQDM | |
| CaMK4 | Q16566 | S12 | TVPSCSASSCSSVTA | + |
| CaMK4 | Q16566 | S13 | VPSCSASSCSSVTAS | + |
| CaMK4 | Q16566 | S336 | AVKAVVASSRLGSAS | - |
| CBP | Q92793 | S302 | PQLASKQSMVNSLPT | |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| gp130 | P40189 | S782 | QVFSRSESTQPLLDS | - |
| HDAC4 | P56524 | S246 | FPLRKTASEPNLKLR | |
| HDAC4 | P56524 | S467 | RPLGRTQSAPLPQNA | |
| HDAC4 | P56524 | S632 | RPLSRAQSSPASATF | |
| HDAC5 | Q9UQL6 | S259 | FPLRKTASEPNLKVR | |
| HDAC5 | Q9UQL6 | S498 | RPLSRTQSSPLPQSP | |
| NFkB-p65 | Q04206 | S536 | SGDEDFSSIADMDFS | |
| NUMB | P49757 | S276 | EQLARQGSFRGFPAL | |
| STMN1 | P16949 | S16 | KELEKRASGQAFELI |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
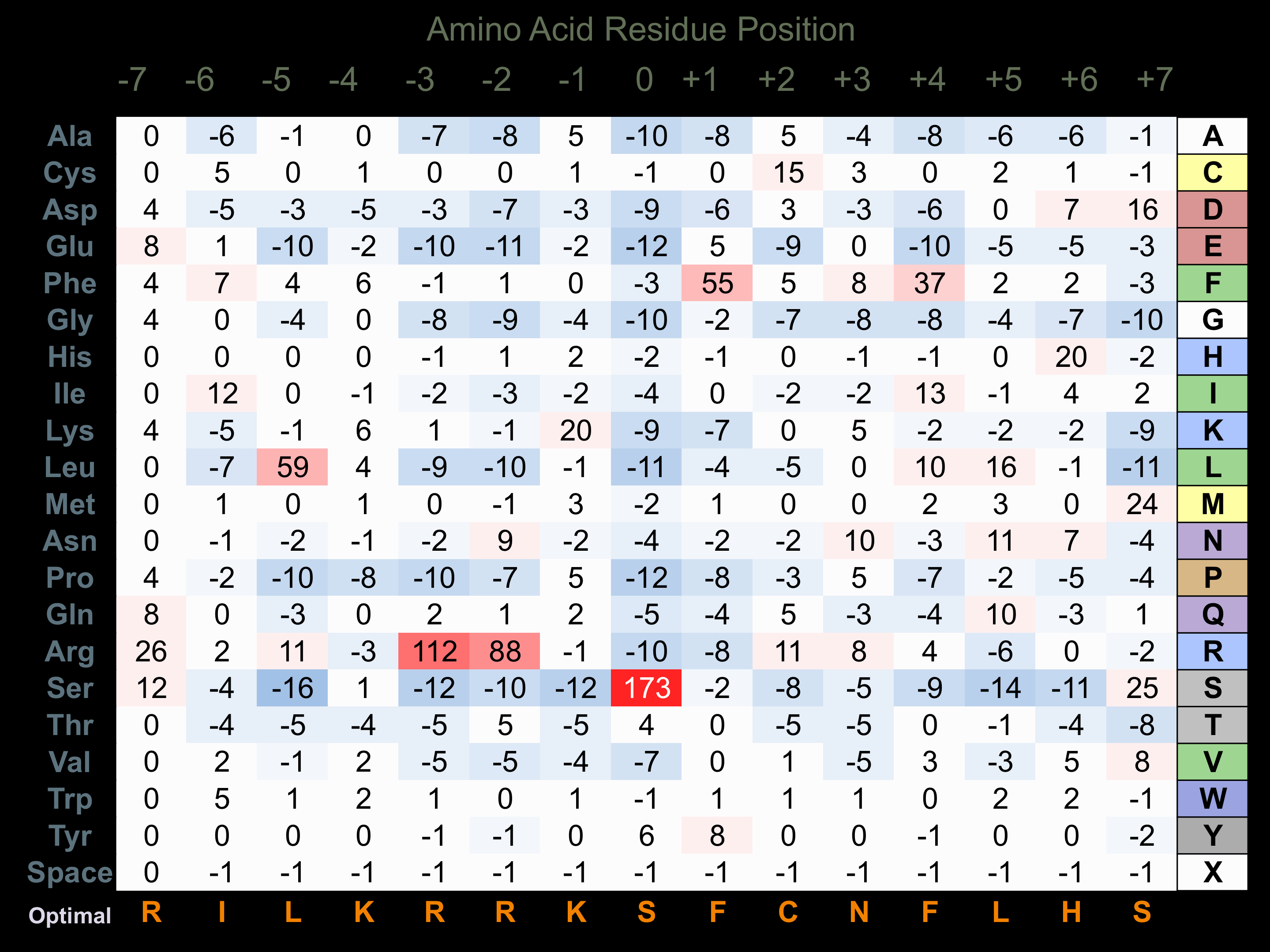
Matrix Type:
Experimentally derived from alignment of 19 known protein substrate phosphosites and 28 peptides phosphorylated by recombinant CAMK4 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Staurosporine | IC50 = 4 nM | 5279 | 8709095 | |
| NVP-TAE684 | Kd = 30 nM | 16038120 | 509032 | 22037378 |
| AT9283 | IC50 > 100 nM | 24905142 | 19143567 | |
| CHEMBL1650545 | IC50 > 100 nM | 53316611 | 1650545 | 21128646 |
| GSK690693 | Kd = 370 nM | 16725726 | 494089 | 22037378 |
| GSK1838705A | Kd = 470 nM | 25182616 | 464552 | 22037378 |
| A674563 | Kd = 710 nM | 11314340 | 379218 | 22037378 |
| Sunitinib | Kd = 890 nM | 5329102 | 535 | 18183025 |
| CP673451 | IC50 > 1 µM | 10158940 | 15705896 | |
| MK5108 | IC50 > 1 µM | 24748204 | 20053775 | |
| Silmitasertib | IC50 > 1 µM | 24748573 | 21174434 | |
| SNS314 | IC50 > 1 µM | 16047143 | 514582 | 18678489 |
| SureCN10063060 | Ki > 1 µM | 52936621 | 21391610 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Momelotinib | IC50 > 2 µM | 25062766 | 19295546 | |
| Lestaurtinib | Kd = 2.7 µM | 126565 | 18183025 | |
| SU14813 | Kd = 2.7 µM | 10138259 | 1721885 | 18183025 |
| Alvocidib | Kd = 3.2 µM | 9910986 | 428690 | 18183025 |
| AST-487 | Kd = 3.7 µM | 11409972 | 574738 | 18183025 |
| Nintedanib | Kd = 3.7 µM | 9809715 | 502835 | 22037378 |
| JNJ-28871063 | IC50 > 4 µM | 17747413 | 17975007 |
Disease Linkage
General Disease Association:
Brain
Specific Diseases (Non-cancerous):
Allan-Herndon-Dudley syndrome; Cerebral hypoxia
Comments:
Genetic or pharmacologic inhibition of CaMK4 in mouse models decreased the frequency of IL-17–producing T cells and ameliorated EAE (an animal model for multiple sclerosis) and lupus-like disease. These studies indicate that CaMK4 has potential as a target for therapeutic inhibition of Th17-driven autoimmune diseases. The Allan-Herndon-Dudley syndrome is an X-linked syndrome of severe intellectual deficit and neurological impairment, and can be caused by deficiencies in T3 levels or mutations of the monocarboxylate transporter 8 gene (MCT8, SLC16A2). T3 normally induces CaMK4 expression in cultured primary neurons.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer stage 2A (%CFC= +132, p<0.062); and T-cell prolymphocytic leukemia (%CFC= +49, p<0.052). The COSMIC website notes an up-regulated expression score for CAMK4 in diverse human cancers of 288, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. Mutations at S12A, S13A, T57A+K58E, K75E, T200A can result in loss of phosphotransferase activity or loss of activation by the upstream kinases CaMKK1 or CaMKK2. Mutations at S189A, H309D+M310E+T312D+F320D+N321D can lead to a constitutively active form.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 24776 diverse cancer specimens. This rate is a modest 1.54-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.76 % in 805 skin cancers tested; 0.56 % in 1093 large intestine cancers tested; 0.28 % in 1669 lung cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,060 cancer specimens
Comments:
Only 5 deletions, 1 insertion and no complex mutations are noted on the COSMIC website.