Nomenclature
Short Name:
CaMKK1
Full Name:
Calcium-calmodulin-dependent protein kinase kinase 1
Alias:
- Calcium/calmodulin-dependent protein kinase 1 alpha, isoform a
- Calcium/calmodulin-dependent protein kinase kinase 1, alpha
- EC 2.7.11.17
- KKCC1
- MGC34095
- CaMKK alpha protein
- CAMKK1.
- CAMKKA
- DKFZP761M0423
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
CAMKK
SubFamily:
Meta
Specific Links
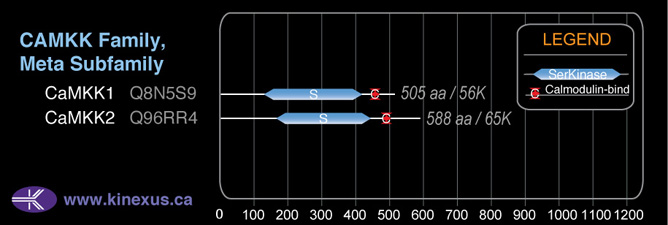
Structure
Mol. Mass (Da):
55,735
# Amino Acids:
505
# mRNA Isoforms:
2
mRNA Isoforms:
57,523 Da (520 AA; Q8N5S9-2); 55,735 Da (505 AA; Q8N5S9)
4D Structure:
Interacts with CAMK4 and calmodulin
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 128 | 409 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
R469, R472.
Serine phosphorylated:
S52, S67, S69, S74+, S97, S100, S443, S452, S458-, S473, S475-, S492.
Threonine phosphorylated:
T108+, T446.
Tyrosine phosphorylated:
Y83.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1427
15
1177
 3
3
38
8
18
 3
3
40
14
37
 13
13
186
69
204
 32
32
457
23
364
 2
2
24
18
23
 1.3
1.3
19
26
12
 2
2
26
23
21
 2
2
23
6
6
 8
8
110
32
63
 1.1
1.1
16
23
21
 35
35
494
35
650
 2
2
34
15
23
 1.3
1.3
19
6
7
 6
6
91
23
125
 3
3
38
10
32
 5
5
66
90
92
 5
5
66
21
50
 7
7
101
39
92
 29
29
413
57
219
 3
3
41
23
29
 2
2
26
23
22
 2
2
34
15
31
 1.5
1.5
21
21
17
 2
2
32
23
35
 34
34
491
53
605
 2
2
23
21
17
 3
3
45
21
29
 3
3
49
21
35
 34
34
484
14
130
 52
52
748
12
61
 63
63
892
16
2079
 1.3
1.3
18
24
19
 62
62
885
52
753
 6
6
87
44
78
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
97 99.5
99.5
99.5
100 -
-
-
94 -
-
-
- 89.5
89.5
91.2
93 -
-
-
- 93.4
93.4
95.8
93.5 93.6
93.6
95.8
94 -
-
-
- 38.4
38.4
46.5
- 69.4
69.4
79.3
76 69.4
69.4
80.1
- 63.5
63.5
76
74 -
-
-
- -
-
-
50.5 -
-
-
- 42.8
42.8
59.7
- 35.7
35.7
50.3
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CALM1 - P62158 |
| 2 | CAMK4 - Q16566 |
| 3 | YWHAH - Q04917 |
| 4 | AKT1 - P31749 |
| 5 | CAMK1 - Q14012 |
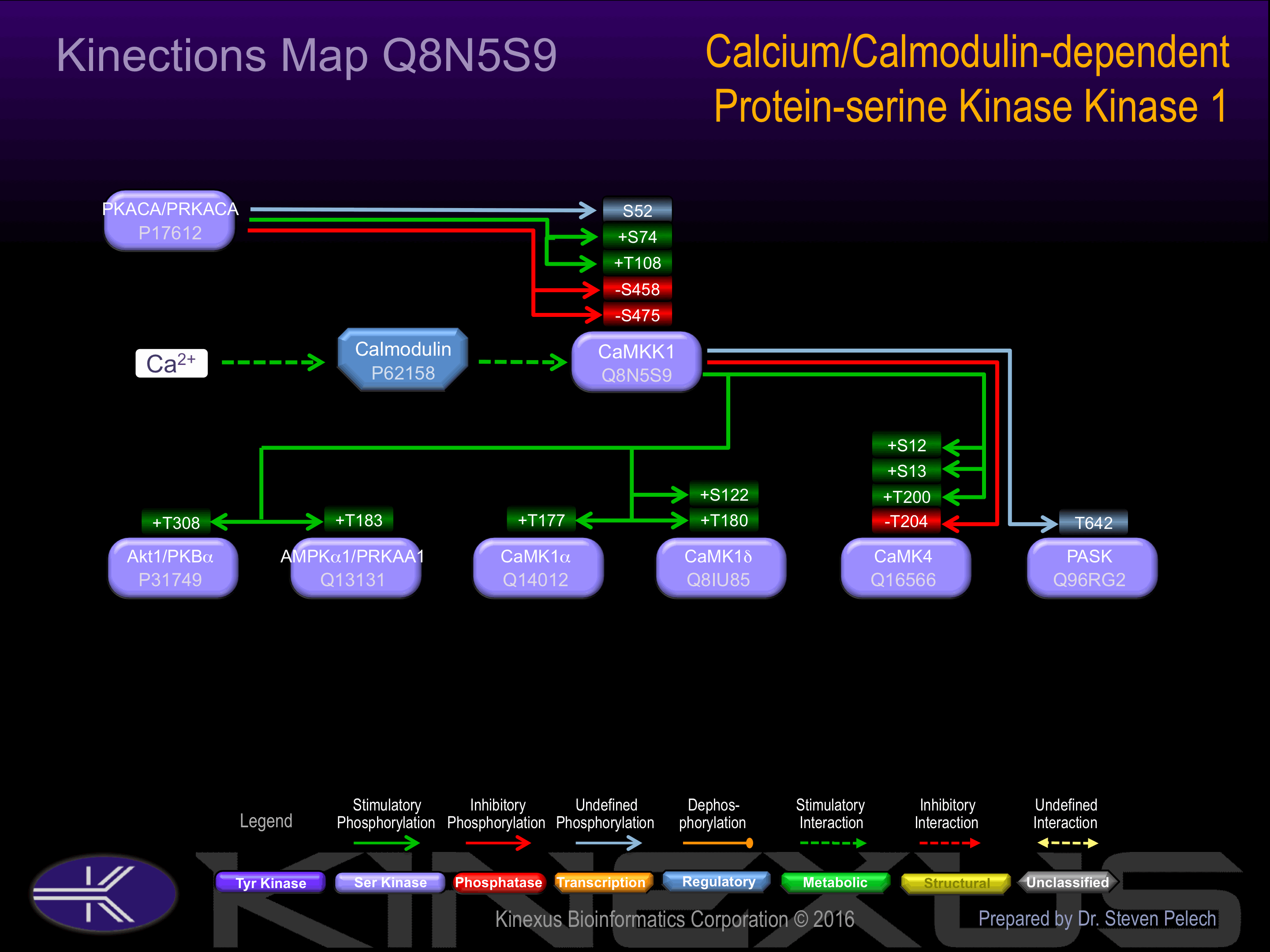
Regulation
Activation:
Activated by Ca2+/calmodulin.
Inhibition:
Binding of calmodulin may releave intrasteric autoinhibition. Partially inhibited upon phosphorylation by PRCAKA/PKA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Akt1 (PKBa) | P31749 | T308 | KDGATMKTFCGTPEY | + |
| AMPKa1 (PRKAA1) | Q13131 | T183 | SDGEFLRTSCGSPNY | + |
| CaMK1a | Q14012 | T177 | DPGSVLSTACGTPGY | + |
| CaMK1d | Q8IU85 | S122 | FYTEKDASTLIRQVL | + |
| CaMK1d | Q8IU85 | T180 | GKGDVMSTACGTPGY | + |
| CaMK4 | Q16566 | S12 | TVPSCSASSCSSVTA | + |
| CaMK4 | Q16566 | S13 | VPSCSASSCSSVTAS | + |
| CaMK4 | Q16566 | T200 | EHQVLMKTVCGTPGY | + |
| CaMK4 | Q16566 | T204 | LMKTVCGTPGYCAPE | - |
| PASK | Q96RG2 | T642 | GLSFGTPTLDEPWLG |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
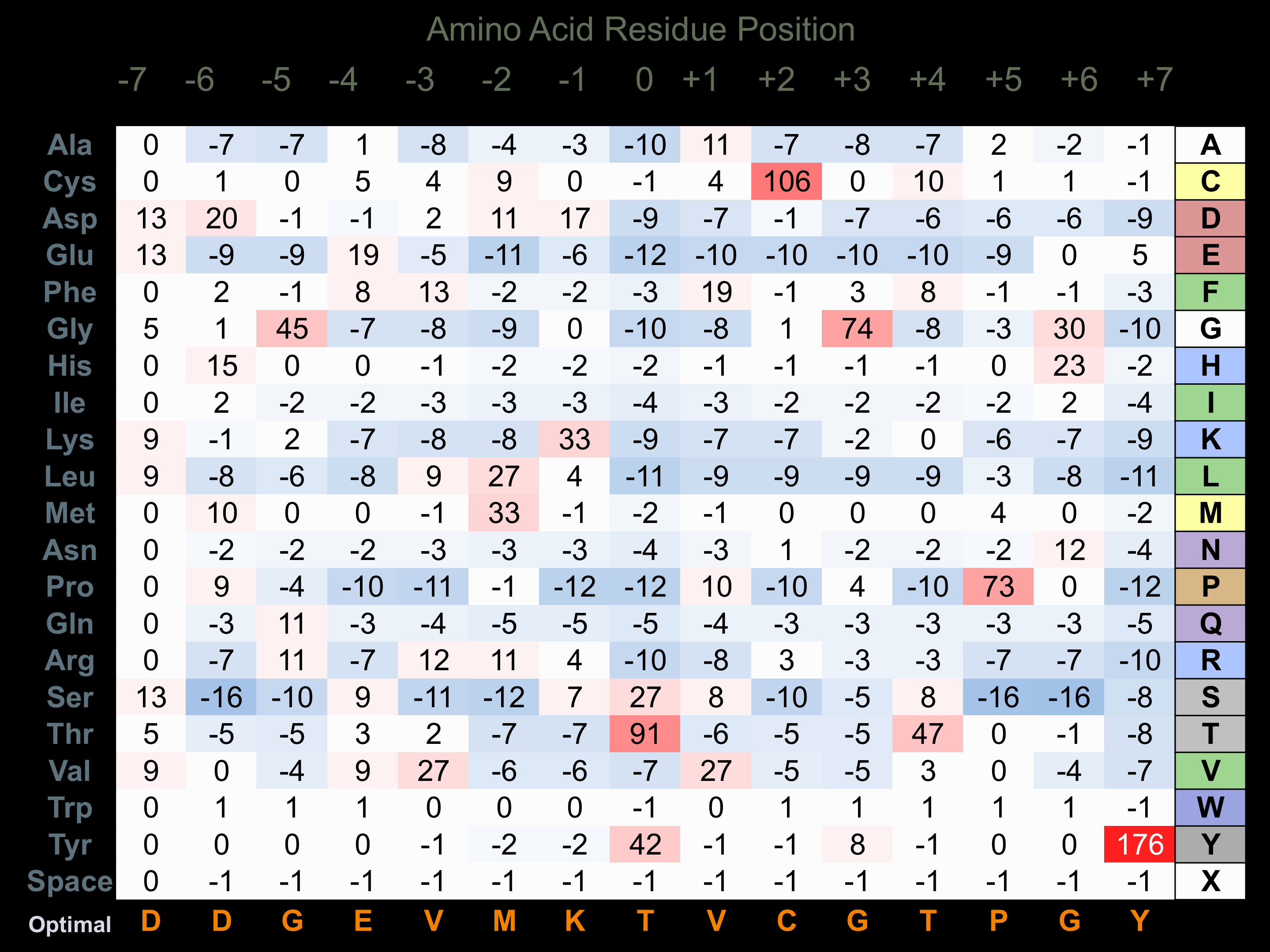
Matrix Type:
Experimentally derived from alignment of 17 known protein substrate phosphosites and 14 peptides phosphorylated by recombinant CaMKK1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Viral infectious disease
Specific Diseases (Non-cancerous):
Cytomegalovirus (CMV) infection
Comments:
CAMKK1 has a role that is important for both cytomegalovirus (CMV) replication and glycolytic activation.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for CaMKK1 in diverse human cancers of 364, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 41 for this protein kinase in human cancers was 0.7-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24726 diverse cancer specimens. This rate is only -10 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.33 % in 603 endometrium cancers tested; 0.3 % in 864 skin cancers tested; 0.3 % in 1270 large intestine cancers tested; 0.24 % in 589 stomach cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,009 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.