Nomenclature
Short Name:
EPHB2
Full Name:
Ephrin type-B receptor 2
Alias:
- CEK5
- EPHB2
- Ephrin type-B receptor 2
- EPHT3
- HEK5; Tyro5
- DRT
- EC 2.7.10.1
- EPH receptor B2
- EPH3
- EPH-3
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Eph
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
117,493
# Amino Acids:
1055
# mRNA Isoforms:
3
mRNA Isoforms:
117,493 Da (1055 AA; P29323); 110,030 Da (987 AA; P29323-3); 109,874 Da (986 AA; P29323-2)
4D Structure:
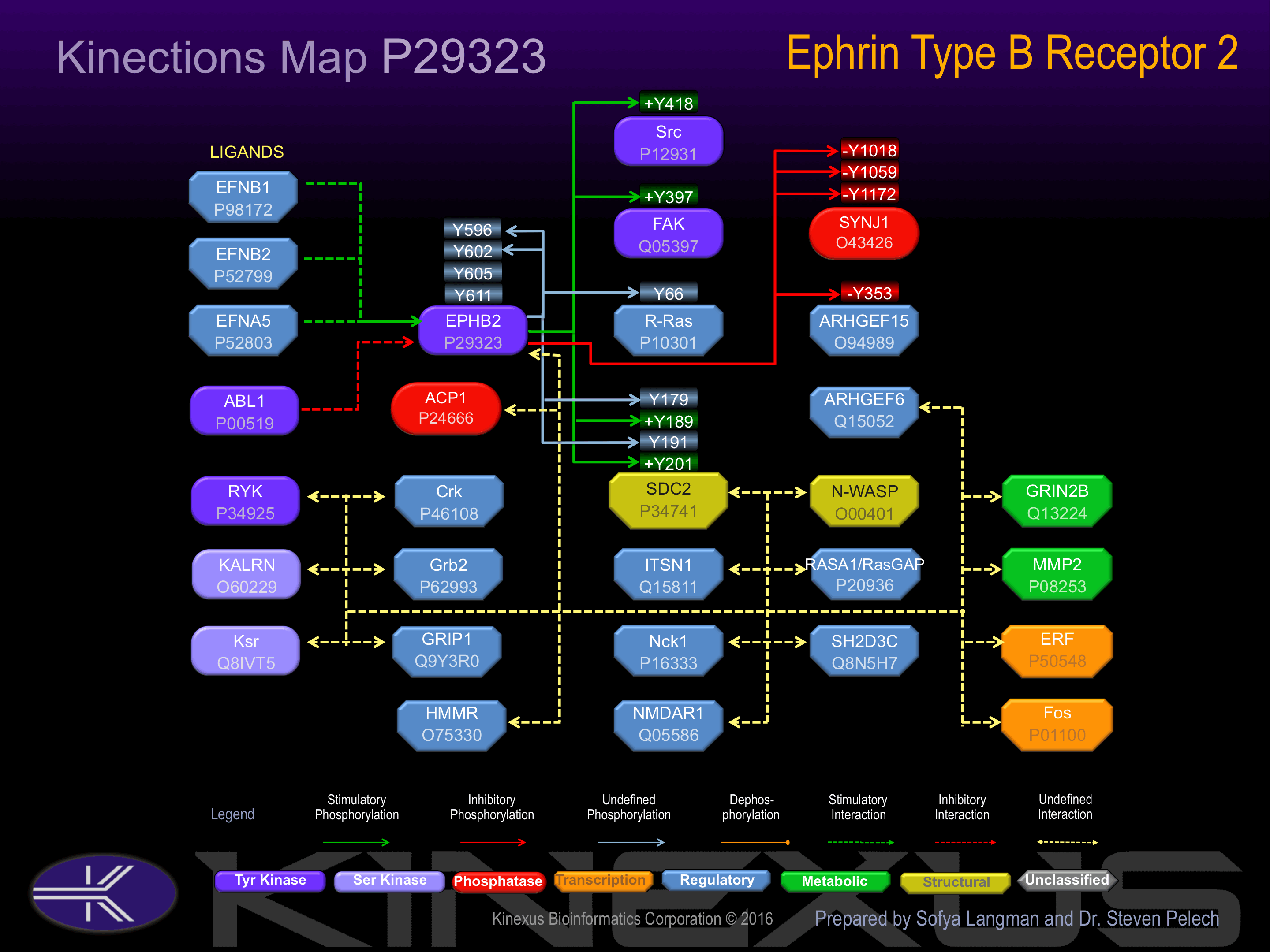
The ligand-activated form interacts with multiple proteins, including GTPase-activating protein (RASGAP) through its SH2 domain. Binds RASGAP through the juxtamembrane tyrosines residues. Interacts with PRKCABP and GRIP1
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
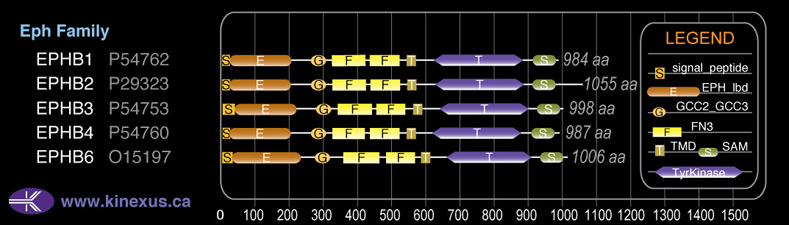
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K137K1020K1024.
N-GlcNAcylated:
N265, N336, N428,N482.
Serine phosphorylated:
S148, S575, S586, S756, S763, S768, S776, S782.
Threonine phosphorylated:
T578, T585, T590, T601, T775, T779, T781, T959.
Tyrosine phosphorylated:
Y175, Y181, Y194, Y481, Y524, Y577, Y584, Y596, Y602, Y742, Y780, Y800, Y912.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 78
78
1002
55
1470
 4
4
46
30
51
 4
4
46
4
43
 23
23
291
169
482
 33
33
426
48
377
 10
10
133
156
512
 25
25
316
66
537
 100
100
1287
64
2392
 32
32
408
34
300
 5
5
65
158
75
 3
3
35
47
40
 45
45
584
327
601
 2
2
32
48
22
 4
4
55
25
44
 4
4
53
35
52
 2
2
29
30
23
 11
11
138
367
1953
 3
3
42
23
49
 3
3
38
160
39
 22
22
286
218
332
 5
5
60
30
52
 3
3
34
37
26
 3
3
34
16
37
 4
4
49
22
56
 3
3
35
32
35
 99
99
1271
97
2271
 2
2
29
54
27
 4
4
46
22
52
 2
2
31
22
33
 13
13
163
56
147
 13
13
163
36
211
 59
59
757
66
929
 0.3
0.3
4
68
2
 54
54
697
109
617
 6
6
76
70
70
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 46.2
46.2
63.7
100 68.9
68.9
81.1
- -
-
-
99 -
-
-
98 90.9
90.9
91.2
100 -
-
-
- 92.5
92.5
93
99.5 68.9
68.9
81.1
99 -
-
-
- -
-
-
- 89
89
91.7
96 66.5
66.5
78.9
89 51.6
51.6
69.2
87 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | EFNB2 - P52799 |
| 2 | EFNB1 - P98172 |
| 3 | MLLT4 - P55196 |
| 4 | AQP1 - P29972 |
| 5 | RRAS - P10301 |
| 6 | VAV2 - P52735 |
| 7 | SDC2 - P34741 |
| 8 | ARHGEF6 - Q15052 |
| 9 | SH2D3C - Q8N5H7 |
| 10 | FAM188B - Q4G0A6 |
| 11 | INMT - O95050 |
| 12 | ERF - P50548 |
| 13 | GRIN2B - Q13224 |
| 14 | ACP1 - P24666 |
| 15 | RYK - P34925 |
Regulation
Activation:
Activated by binding ephrin-B1, B2, or B3.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
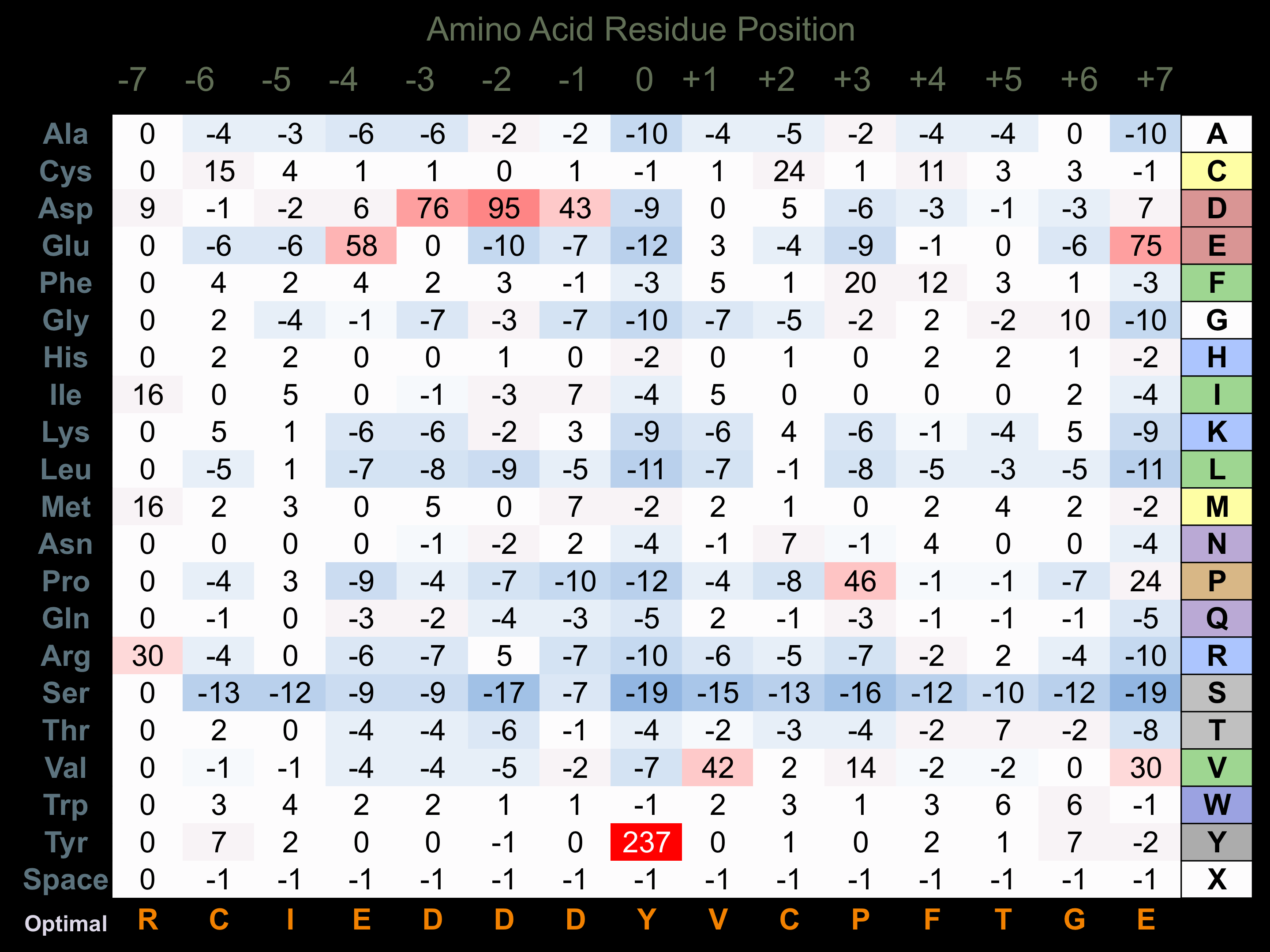
Matrix Type:
Experimentally derived from alignment of 9 known protein substrate phosphosites and 50 peptides phosphorylated by recombinant EphB2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological, and bone disorders
Specific Diseases (Non-cancerous):
Alzheimer's disease; Craniofrontonasal syndrome
Comments:
Eph receptors regulate synaptic localization and/or retention of the NMDA receptor (NMDAR) in mouse brain via direct interaction and through Src-mediated tyrosine phosphorylation.This relationship between EphB2 and NMDAR may underlie the development of Alzheimer’s disease, since Amyloid-beta protein (A beta) binding to EphB2 prevents EphB2-NMDAR interactions, which leads to NMDAR internalization, decreased synaptic strength and memory impairment. Stress-induced cleavage of EphB2 by Neuropsin causes dissociation of EphB2 from the NMDAR subunit NR1, which can promote increased turnover of EphB2. With increased EphB2-NR1 binding, this can enhance NMDAR activity and promote the development of anxiety disorders. In response to EphB2 down-regulation in mice infect with lentivirus that express anti-EphB2 short hairpinRNA, hippocampal granule cells showed declines in long term potentiation, which indicates a role for EphB2 in synaptic plasticity.
Specific Cancer Types:
Familial prostate cancer (FPC); Prostate cancer (PC); Colorectal cancer (CRC)
Comments:
EPHB2 appears to be a tumour suppressor protein (TSP). The active form of the protein kinase normally acts to inhibit tumour cell proliferation. EphB2 mutations have been found in prostate cancer. Most prostate cancers are adenocarcinomas that derived from acini of the prostatic ducts. K1019X mutation was significantly more common in African American probands. Over-expression of EphB2 has been frequently detected in both well-differentiated adenocarcinomas (10 of 13) and poorly differentiated adenocarcinomas (9 of 14).
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer stage 2A (%CFC= +85, p<0.002); Colon mucosal cell adenomas (%CFC= +156, p<0.0001); Gastric cancer (%CFC= +462, p<0.0001); Lung adenocarcinomas (%CFC= +111, p<0.0001); and Ovary adenocarcinomas (%CFC= +103, p<0.003). The COSMIC website notes an up-regulated expression score for EPHB2 in diverse human cancers of 402, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 2 for this protein kinase in human cancers was 97% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 25397 diverse cancer specimens. This rate is only 34 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.69 % in 854 skin cancers tested; 0.34 % in 1093 large intestine cancers tested; 0.33 % in 602 endometrium cancers tested; 0.29 % in 589 stomach cancers tested; 0.14 % in 1942 lung cancers tested; 0.12 % in 1226 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: E615K (5).
Comments:
Only 11 deletions (8 at I393fs*19), and no insertions or complex mutations are noted on the COSMIC website.