Nomenclature
Short Name:
ERK2
Full Name:
Mitogen-activated protein kinase 1
Alias:
- EC 2.7.11.24
- MAP kinase 2
- MAP kinase isoform p42
- MAPK
- MAPK 2; PRKM2
- ERK
- ERK-2
- Extracellular signal regulated kinase 2
- ERT1
- Kinase ERK2
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
MAPK
SubFamily:
ERK
Specific Links
Structure
Mol. Mass (Da):
43,390
# Amino Acids:
360
# mRNA Isoforms:
2
mRNA Isoforms:
41,390 Da (360 AA; P28482); 36,432 Da (316 AA; P28482-2)
4D Structure:
Interacts with MORG1 By similarity. Binds to HIV-1 Nef through its SH3 domain. This interaction inhibits its tyrosine-kinase activity. Interacts with its substrates HSF4 and ARHGEF2. Interacts (phosphorylated form) with CAV2 (Tyr-19-phosphorylated form); the interaction, promoted by insulin, leads to nuclear location and MAPK1 activation By similarity. Interacts with NISCH. Interacts with ARRB2. Interacts with ADAM15.
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 25 | 313 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S29+, S202, S246, S248, S284, S360.
Threonine phosphorylated:
T63, T181+, T185+, T190-, T206, T295.
Tyrosine phosphorylated:
Y25, Y36-, Y43, Y113, Y187+, Y193-, Y205, Y263.
Ubiquitinated:
K55, K99, K203, K292, K344.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1489
56
1082
 8
8
114
20
160
 17
17
249
10
134
 53
53
790
220
1252
 62
62
919
64
609
 46
46
679
142
1656
 13
13
187
81
273
 42
42
621
51
866
 42
42
627
17
517
 17
17
256
157
407
 8
8
116
33
101
 59
59
883
184
690
 7
7
109
32
77
 5
5
71
12
69
 10
10
153
26
164
 9
9
136
40
194
 24
24
355
282
2670
 12
12
186
19
177
 14
14
211
153
221
 54
54
811
239
657
 18
18
265
25
246
 15
15
222
29
153
 9
9
129
20
131
 19
19
276
19
165
 16
16
234
25
222
 50
50
746
142
686
 11
11
168
35
125
 11
11
166
19
137
 9
9
131
19
114
 20
20
304
70
199
 66
66
981
36
557
 29
29
428
61
505
 28
28
423
170
686
 77
77
1142
187
799
 17
17
248
118
255
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 78.1
78.1
85.5
100 -
-
-
100 -
-
-
100 -
-
-
100 96
96
96
100 -
-
-
- 83.4
83.4
89.2
99 99.2
99.2
99.4
99 -
-
-
- -
-
-
- 97.3
97.3
97.5
99 95.8
95.8
97.2
98 92.1
92.1
94.6
95 -
-
-
- 67.8
67.8
74.7
82 81.9
81.9
86.8
- 64.4
64.4
71
81 79.4
79.4
84.5
- -
-
-
- -
-
-
- -
-
-
53 48.7
48.7
65.4
52.5 49.7
49.7
66.4
54 -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
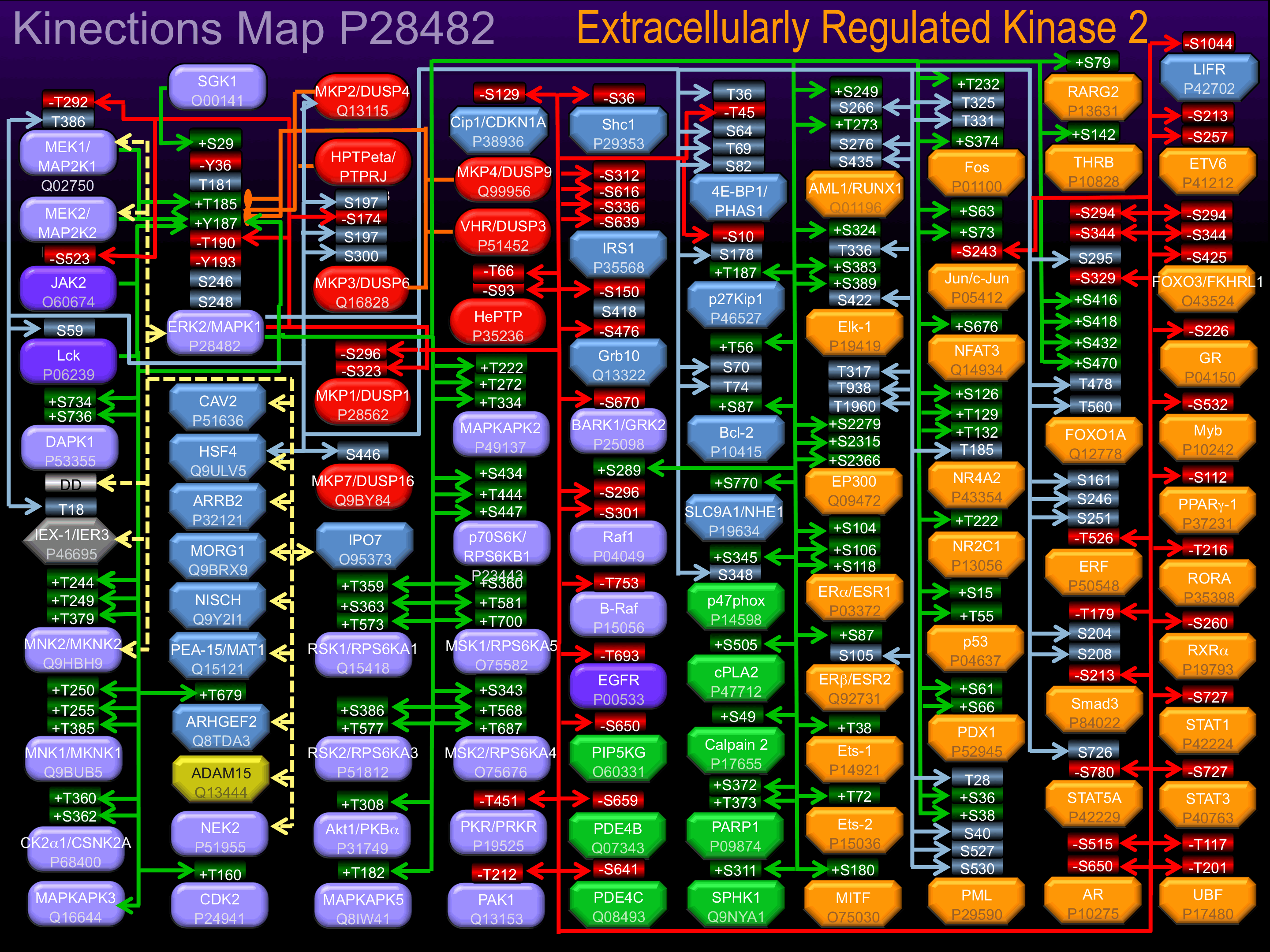
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAP2K1 - Q02750 |
| 2 | RPS6KA1 - Q15418 |
| 3 | MAPK14 - Q16539 |
| 4 | MYC - P01106 |
| 5 | DUSP1 - P28562 |
| 6 | PTPN7 - P35236 |
| 7 | RPS6KA3 - P51812 |
| 8 | MKNK2 - Q9HBH9 |
| 9 | MAP2K2 - P36507 |
| 10 | TH - P07101 |
| 11 | DUSP6 - Q16828 |
| 12 | TP53 - P04637 |
| 13 | TOB1 - P50616 |
| 14 | ELK1 - P19419 |
| 15 | PEA15 - Q15121 |
Regulation
Activation:
Phosphorylation of Thr-185 and Tyr-187 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 1-Cys PRX | P30041 | T176 | TAEKRVATPVDWKDG | |
| 4E-BP1 | Q13541 | S64 | FLMECRNSPVTKTPP | |
| 4E-BP1 | Q13541 | S82 | PTIPGVTSPSSDEPP | |
| 4E-BP1 | Q13541 | T36 | PPGDYSTTPGGTLFS | |
| 4E-BP1 | Q13541 | T45 | GGTLFSTTPGGTRII | - |
| 4E-BP1 | Q13541 | T69 | RNSPVTKTPPRDLPT | |
| 5-LO | P09917 | S663 | QLPYYYLSPDRIPNS | |
| ADAM17 | P78536 | T735 | KPFPAPQTPGRLQPA | |
| AEBP1 | Q8IUX7 | T1012 | IDPSRPMTPQQRRLQ | |
| AML1 (RUNX1) | Q01196 | S249 | DTRQIQPSPPWSYDQ | + |
| AML1 (RUNX1) | Q01196 | S266 | QYLGSIASPSVHPAT | |
| AML1 (RUNX1) | Q01196 | S276 | VHPATPISPGRASGM | |
| AML1 (RUNX1) | Q01196 | S435 | AEGSHSNSPTNMAPS | |
| AML1 (RUNX1) | Q01196 | T273 | SPSVHPATPISPGRA | + |
| Amphiphysin | P49418 | S285 | NHILAPASPAPARPR | |
| Amphiphysin | P49418 | S293 | PAPARPRSPSQTRKG | |
| AR | P10275 | S515 | VSRVPYPSPTCVKSE | - |
| AR | P10275 | S650 | GEASSTTSPTEETTQ | - |
| ARHGAP26 | Q9UNA1 | S685 | PMFSAPSSPMPTSST | |
| ARHGEF2 | Q8TDA3 | T679 | PGVELLLTPREPALP | + |
| ARRB1 | P49407 | S412 | EEEDGTGSPQLNNR_ | |
| ASC-2 | Q14686 | S884 | NKDVTLTSPLLVNLL | |
| ATF2 | P15336 | T69 | SVIVADQTPTPTRFL | + |
| ATF2 | P15336 | T71 | IVADQTPTPTRFLKN | + |
| ATF7 | P17544 | T51 | SVIIADQTPTPTRFL | - |
| ATF7 | P17544 | T53 | IIADQTPTPTRFLKN | - |
| B-Raf | P15056 | T753 | YACASPKTPIQAGGY | - |
| Bcl-2 | P10415 | S70 | RDPVARTSPLQTPAA | |
| Bcl-2 | P10415 | S87 | AAAGPALSPVPPVVH | + |
| Bcl-2 | P10415 | T56 | FSSQPGHTPHPAASR | + |
| Bcl-2 | P10415 | T74 | ARTSPLQTPAAPGAA | |
| Bcl-6 | P41182 | S333 | KGLVSPQSPQKSDCQ | |
| Bcl-6 | P41182 | S343 | KSDCQPNSPTESCSS | |
| Bim | O43521 | S104 | FSFDTDRSPAPMSCD | |
| Bim | O43521 | S113 | APMSCDKSTQTPSPP | |
| Bim | O43521 | S59 | GDSCPHGSPQGPLAP | |
| Bim | O43521 | S69 | GPLAPPASPGPFATR | |
| Bim | O43521 | T114 | PMSCDKSTQTPSPPC | |
| BLVRA (BVA) | P53004 | S230 | LKRNRYLSFHFKSGS | |
| BMAL1 | O00327 | T484 | SPLNITSTPPPDASS | |
| BTG2 | P78543 | S147 | QVLLGRSSPSKNYVM | |
| C-EBPa | P49715 | S21 | PMSSHLQSPPHAPSS | |
| C-EBPb | P17676 | T235 | SSSSPPGTPSPADAK | |
| CACNA1B | Q00975 | S411 | DRNAEEKSPLDVLKR | |
| CACNA1B | Q00975 | S446 | ADLCAVGSPFARASL | |
| CACNB1 | Q02641 | S161 | CEVGFIPSPVKLDSL | |
| CACNB1 | Q02641 | S348 | IVYIKITSPKVLQRL | |
| CAD | P27708 | T456 | KVYFLPITPHYVTQV | |
| Caldesmon | Q05682 | S759 | KTPDGNKSPAPKPSD | |
| Caldesmon | Q05682 | S789 | QSVDKVTSPTKV___ | |
| Calpain 2 | P17655 | S49 | GTLFQDPSFPAIPSA | + |
| Caspase 9 | P55211 | T125 | PEVLRPETPRPVDIG | |
| CDK2 | P24941 | T160 | GVPVRTYTHEVVTLW | + |
| CENPE | Q02224 | S2605 | KQVTCENSPKSPKVT | |
| CENPE | Q02224 | S2608 | TCENSPKSPKVTGTA | |
| CENPE | Q02224 | S2639 | QDPVPKESPKSCFFD | |
| CENPE | Q02224 | S2654 | SRSKSLPSPHPVRYF | |
| CEP55 | Q53EZ4 | S425 | NREKVAASPKSPTAA | |
| CEP55 | Q53EZ4 | S428 | KVAASPKSPTAALNE | |
| CIITA | P33076 | S280 | TVHGLPTSPDRPGST | |
| CIITA | P33076 | S288 | PDRPGSTSPFAPSAT | |
| Cip1 (p21, CDKN1A) | P38936 | S129 | SGEQAEGSPGGPGDS | - |
| Cip1 (p21, CDKN1A) | P38936 | T56 | NFDFVTETPLEGDFA | |
| CK2a1 (CSNK2A1) | P68400 | S362 | ISSVPTPSPLGPLAG | + |
| CK2a1 (CSNK2A1) | P68400 | T360 | SGISSVPTPSPLGPL | + |
| cPLA2 | P47712 | S505 | LNTSYPLSPLSDFAT | + |
| CRY1 | Q16526 | S247 | NANSLLASPTGLSPY | |
| CRY2 | Q8IV71 | S266 | NANSLLASPTGLSPY | |
| CRYAB | P02511 | S45 | FPTSTSLSPFYLRPP | |
| CRYAB | P02511 | S59 | PSFLRAPSWFDTGLS | ? |
| CTTN (Cortactin) | Q14247 | S405 | KTQTPPVSPAPQPTE | - |
| CTTN (Cortactin) | Q14247 | S418 | TEERLPSSPVYEDAA | |
| Cx43 | P17302 | S254 | HATSGALSPAKDCGS | |
| Cx43 | P17302 | S278 | SSPTAPLSPMSPPGY | |
| Cx43 | P17302 | S281 | TAPLSPMSPPGYKLV | |
| Cyclin B1 | P14635 | S126 | PILVDTASPSPMETS | |
| Cyclin B1 | P14635 | S128 | LVDTASPSPMETSGC | |
| DAPK1 | P53355 | S734 | NSSRFPPSPLASKPT | + |
| DAPK1 | P53355 | S736 | NSSRFPPSPLASKPT | + |
| DAT (Dopamine transporter) | Q01959 | S53 | TLTNPRQSPVEAQDR | |
| DAZAP1 | Q96EP5 | T269 | FTSYIVSTPPGGFPP | |
| DAZAP1 | Q96EP5 | T315 | GVPPPPATPGAAPLA | |
| DDHD1 | Q8NEL9 | S727 | TIPSPVTSPVLSRRH | |
| DLG4 (PSD-95) | P78352 | S290 | PTAMTPTSPRRYSPV | |
| DLG4 (PSD-95) | P78352 | T287 | TDYPTAMTPTSPRRY | |
| DSCR1 | P53805 | S112 | FLISPPASPPVGWKQ | |
| DUSP1 (MPK1) | P28562 | S296 | KQRRSIISPNFSFMG | - |
| DUSP1 (MPK1) | P28562 | S323 | HCSAEAGSPAMAVLD | - |
| E2A | P15923 | S352 | SSNNFSSSPSTPVGS | |
| E2A | P15923 | S359 | SPSTPVGSPQGLAGT | |
| E2A | P15923 | T355 | NFSSSPSTPVGSPQG | |
| EGFR | P00533 | T693 | RELVEPLTPSGEAPN | - |
| eIF2A | P05198 | S52 | MILLSELSRRRIRSI | - |
| eIF2B-e | Q13144 | S544 | EPDSRGGSPQMDDIK | |
| Elk-1 | P19419 | S324 | RDLELPLSPSLLGGP | + |
| Elk-1 | P19419 | S324 | RDLELPLSPSLLGGP | + |
| Elk-1 | P19419 | S383 | IHFWSTLSPIAPRSP | + |
| Elk-1 | P19419 | S389 | LSPIAPRSPAKLSFQ | + |
| Elk-1 | P19419 | S422 | LSTPVVLSPGPQKP_ | + |
| Elk-1 | P19419 | T336 | GGPGPERTPGSGSGS | |
| Elk-1 | P19419 | T353 | QAPGPALTPSLLPTH | + |
| Elk-1 | P19419 | T363 | LLPTHTLTPVLLTPS | + |
| Elk-1 | P19419 | T368 | TLTPVLLTPSSLPPS | + |
| Elk-1 | P19419 | T417 | ISVDGLSTPVVLSPG | + |
| ENaC beta | P51168 | T615 | QALPIPGTPPPNYDS | |
| ENaC gamma | P51170 | T622 | LGTQVPGTPPPKYNT | |
| EP300 | Q09472 | S2279 | PVQPNPMSPQQHMLP | + |
| EP300 | Q09472 | S2315 | RSPQPVPSPRPQSQP | + |
| EP300 | Q09472 | S2366 | MEQGHFASPDQNSML | + |
| EP300 | Q09472 | T1960 | QHQMPPMTPMAPMGM | |
| EP300 | Q09472 | T317 | VQQPGLVTPVAQGMG | |
| EP300 | Q09472 | T938 | LLPPQPATPLSQPAV | |
| ERa (ESR1) | P03372 | S104 | FPPLNSVSPSPLMLL | + |
| ERa (ESR1) | P03372 | S106 | PLNSVSPSPLMLLHP | + |
| ERa (ESR1) | P03372 | S118 | LHPPPQLSPFLQPHG | + |
| ERb | Q92731 | S105 | LYAEPQKSPWCEARS | + |
| ERb | Q92731 | S87 | WPTPGHLSPLVVHRQ | + |
| ERF | P50548 | S161 | SPTEDPRSPPACSSS | ? |
| ERF | P50548 | S246 | RGGPEPLSPFPVSPL | ? |
| ERF | P50548 | S251 | PLSPFPVSPLAGPGS | ? |
| ERF | P50548 | T526 | GEAGGPLTPRRVSSD | - |
| Ets-1 | P14921 | T38 | CADVPLLTPSSKEMM | + |
| Ets-2 | P15036 | T72 | NCELPLLTPCSKAVM | + |
| ETV1 (ER81) | P50549 | S146 | TPSSTPVSPLHHASP | |
| ETV1 (ER81) | P50549 | S94 | KIKKEPHSPCSEISS | |
| EWS | Q01844 | T79 | QPPTGYTTPTAPQAY | |
| FAK (PTK2) | Q05397 | S910 | KLQPQEISPPPTANL | ? |
| Fe65 | O00213 | S175 | EEEEDLSSPPGLPEP | |
| Fe65 | O00213 | S287 | WEPPGRASPSQGSSP | |
| Fe65 | O00213 | S347 | TFPAQSLSPEPLPQE | |
| Fe65 | O00213 | T709 | PKRLGAHTP______ | |
| Fos | P01100 | S374 | PSSDSLSSPTLLAL_ | + |
| Fos | P01100 | T232 | GGLPEVATPESEEAF | + |
| Fos | P01100 | T325 | TELEPLCTPVVTCTP | |
| Fos | P01100 | T331 | CTPVVTCTPSCTAYT | |
| FOXO1A | Q12778 | S249 | EGGKSGKSPRRRAAS | - |
| FOXO1A | Q12778 | S287 | GQEGAGDSPGSQFSK | - |
| FOXO1A | Q12778 | S295 | QFSKWPASPGSHSND | |
| FOXO1A | Q12778 | S329 | STISGRLSPIMTEQD | - |
| FOXO1A | Q12778 | S416 | PPNTSLNSPSPNYQK | + |
| FOXO1A | Q12778 | S418 | NTSLNSPSPNYQKYT | + |
| FOXO1A | Q12778 | S432 | TYGQSSMSPLPQMPI | + |
| FOXO1A | Q12778 | S470 | KELLTSDSPPHNDIM | + |
| FOXO1A | Q12778 | T478 | PPHNDIMTPVDPGVA | |
| FOXO1A | Q12778 | T560 | NRLTQVKTPVQVPLP | |
| FOXO3 (FKHRL1) | O43524 | S294 | QLSKWPGSPTSRSSD | - |
| FOXO3 (FKHRL1) | O43524 | S344 | QDDDAPLSPMLYSSS | - |
| FOXO3 (FKHRL1) | O43524 | S425 | TKGSGLGSPTSSFNS | - |
| GAB1 | Q13480 | T476 | EANYVPMTPGTFDFS | |
| GAB2 | Q9UQC2 | S623 | ALDFQPSSPSPHRKP | |
| GABRA1 | P14867 | T376 | APTATSYTPNLARGD | |
| GAIP | P49795 | S151 | EDYVSILSPKEVSLD | |
| GAP43 | P17677 | S151 | TKASTDNSPSSKAED | |
| GATA1 | P15976 | S178 | TFFSPTGSPLNSAAY | |
| GATA1 | P15976 | S26 | VDPALVSSTPESGVF | |
| GATA4 | P43694 | S105 | AYTPPPVSPRFSFPG | |
| GATA6 | Q92908 | S266 | VSARFPYSPSPPMAN | |
| GORASP2 | Q9H8Y8 | T222 | LPGQMAGTPITPLKD | |
| GR | P04150 | S226 | IDENCLLSPLAGEDD | - |
| GRASP65 | Q9BQQ3 | S273 | DPLPGPGSPSHSAPD | |
| Grb10 | Q13322 | S150 | PELCGPGSPPVLTPG | - |
| Grb10 | Q13322 | S418 | QQRKALLSPFSTPVR | |
| Grb10 | Q13322 | S476 | MNILGSQSPLHPSTL | - |
| GSK3b | P49841 | T43 | KVTTVVATPGQGPDR | |
| GTF2I | P78347 | S668 | INTKALQSPKRPRSP | |
| GTF2I | P78347 | S674 | QSPKRPRSPGSNSKV | |
| H3.2 | P84228 | S29 | ATKAARKSAPATGGV | |
| HePTP | P35236 | S93 | ALQRQPPSPKQLEEE | - |
| HePTP | P35236 | T66 | EPICSVNTPREVTLH | - |
| HIF1A | Q16665 | S641 | DIKILIASPSPTHIH | |
| HIF1A | Q16665 | S643 | KILIASPSPTHIHKE | |
| hnRNP K | P61978 | S284 | RRDYDDMSPRRGPPP | |
| hnRNP K | P61978 | S353 | DSAIDTWSPSEWQMA | |
| HSF1 | Q00613 | S303 | RVKEEPPSPPQSPRV | - |
| HSF1 | Q00613 | S307 | EPPSPPQSPRVEEAS | - |
| IEX-1 (IER3) | P46695 | T18 | MTILQAPTPAPSTIP | |
| ILKAP | Q9H0C8 | T315 | DIRRCQLTPNDRFIL | |
| ILKAP | Q9H0C8 | T333 | DGLFKVFTPEEAVNF | |
| iNOS | P35228 | S745 | KSRQNLQSPTSSRAT | |
| IP3R1 (IP3 receptor) | Q14643 | S1774 | SFGNGPLSAGGPGKP | |
| IP3R1 (IP3 receptor) | Q14643 | S436 | AFAIVPVSPAEVRDL | |
| IRS1 | P35568 | S312 | TESITATSPASMVGG | - |
| IRS1 | P35568 | S616 | DDGYMPMSPGVAPVP | - |
| IRS1 | P35568 | S636 | SGDYMPMSPKSVSAP | - |
| IRS1 | P35568 | S639 | YMPMSPKSVSAPQQI | - |
| Irx2 | Q9BZI1 | S317 | TPQGSRTSPGAPPPA | |
| Irx2 | Q9BZI1 | S46 | SASGSAFSPYPGSAA | |
| Irx2 | Q9BZI1 | S64 | QAATGFGSPLQYSAD | |
| JAK2 | O60674 | S523 | GVSDVPTSPTLQRPT | - |
| Jun (c-Jun) | P05412 | S243 | PGETPPLSPIDMESQ | - |
| Jun (c-Jun) | P05412 | S63 | KNSDLLTSPDVGLLK | + |
| Jun (c-Jun) | P05412 | S73 | VGLLKLASPELERLI | + |
| JunD | P17535 | S100 | LGLLKLASPELERLI | + |
| JunD | P17535 | S90 | PPDGLLASPDLGLLK | + |
| Kir6.2 | Q14654 | S385 | AKPKFSISPDSLS__ | |
| Kir6.2 | Q14654 | T341 | GNTIKVPTPLCTARQ | |
| KRT8 | P05787 | S432 | SAYGGLTSPGLSYSL | |
| KRT8 | P05787 | S74 | TVNQSLLSPLVLEVD | |
| Ksr1 (KSR) | Q8IVT5 | S443 | AMNHLDSSSNPSSTT | |
| Ksr1 (KSR) | Q8IVT5 | S456 | TTSSTPSSPAPFPTS | |
| Ksr1 (KSR) | Q8IVT5 | T272 | SFITPPTTPQLRRHT | |
| Ksr1 (KSR) | Q8IVT5 | T286 | TKLKPPRTPPPPSRK | |
| Kv4.2 | Q9NZV8 | S616 | EGDDRPESPEYSGGN | |
| Kv4.2 | Q9NZV8 | T602 | TAIISIPTPPVTTPE | |
| Kv4.2 | Q9NZV8 | T607 | IPTPPVTTPEGDDRP | |
| Lck | P06239 | S59 | EGSNPPASPLQDNLV | ? |
| LIFR | P42702 | S1044 | WNLVSPDSPRSIDSN | - |
| MAP2 | P11137 | T1616 | YSSRTPGTPGTPSYP | |
| MAP2 | P11137 | T1619 | RTPGTPGTPSYPRTP | |
| MAPKAPK2 | P49137 | S272 | SNHGLAISPGMKTRI | + |
| MAPKAPK2 | P49137 | S9 | LSNSQGQSPPVPFPA | |
| MAPKAPK2 | P49137 | T222 | TSHNSLTTPCYTPYY | + |
| MAPKAPK2 | P49137 | T25 | APPPQPPTPALPHPP | |
| MAPKAPK2 | P49137 | T334 | QSTKVPQTPLHTSRV | + |
| MAPKAPK2 | P49137 | Y367 | LATMRVDYEQIKIKK | |
| MAPKAPK2 | P49137 | Y63 | KNAIIDDYKVTSQVL | |
| MAPKAPK5 | Q8IW41 | T182 | IDQGDLMTPQFTPYY | + |
| MAZ | P56270 | T72 | AAPAPPPTPQAPAAE | |
| MBP | P02686 | T232 | KNIVTPRTPPPSQGK | |
| MCL1 | Q07820 | T163 | TDGSLPSTPPPAEEE | |
| MEK1 (MAP2K1) | Q02750 | T292 | ETPPRPRTPGRPLSS | - |
| MEK1 (MAP2K1) | Q02750 | T386 | IGLNQPSTPTHAAGV | ? |
| MITF | O75030 | S180 | PGSSAPNSPMAMLTL | + |
| MKL1 | Q969V6 | S454 | TGSTPPVSPTPSERS | |
| MKP3 (DUSP6) | Q16828 | S159 | DGSCSSSSPPLPVLG | |
| MKP3 (DUSP6) | Q16828 | S197 | SATDSDGSPLSNSQP | |
| MKP7 (DUSP16) | Q9BY84 | S446 | TNKLCQFSPVQELSE | |
| MNK1 (MKNK1) | Q9BUB5 | S226 | CESPEKVSPVKICDF | |
| MNK1 (MKNK1) | Q9BUB5 | T250 | NSCTPITTPELTTPC | + |
| MNK1 (MKNK1) | Q9BUB5 | T255 | ITTPELTTPCGSAEY | + |
| MNK1 (MKNK1) | Q9BUB5 | T385 | APEKGLPTPQVLQRN | + |
| MNK2 (GPRK7) | Q9HBH9 | T244 | GDCSPISTPELLTPC | + |
| MNK2 (GPRK7) | Q9HBH9 | T249 | ISTPELLTPCGSAEY | + |
| MNK2 (GPRK7) | Q9HBH9 | T379 | APENTLPTPMVLQRN | + |
| MSK1 (RPS6KA5) | O75582 | S360 | TEMDPTYSPAALPQS | + |
| MSK1 (RPS6KA5) | O75582 | T581 | PDNQPLKTPCFTLHY | + |
| MSK1 (RPS6KA5) | O75582 | T700 | LSSNPLMTPDILGSS | + |
| MSK2 (RPS6KA4) | O75676 | S343 | TRLEPVYSPPGSPPP | + |
| MSK2 (RPS6KA4) | O75676 | T568 | SPGVPMQTPCFTLQY | + |
| MSK2 (RPS6KA4) | O75676 | T687 | RSSPPLRTPDVLESS | + |
| Myc | P01106 | S62 | LLPTPPLSPSRRSGL | |
| Myc | P01106 | T58 | KKFELLPTPPLSPSR | |
| MYLK1 (smMLCK) | Q15746 | S1005 | GLSGRKSSTGSPTSP | |
| MYLK1 (smMLCK) | Q15746 | S1779 | SSTGSPTSPLNAEKL | |
| N-CAM L1 | P32004 | S1204 | GDIKPLGSDDSLADY | |
| N-CAM L1 | P32004 | S1248 | NDSSGATSPINPAVA | |
| NCOA1 | Q15788 | S1185 | GTPPASTSPFSQLAA | |
| NCOA1 | Q15788 | S395 | PSVNPSISPAHGVAR | |
| NCOA1 | Q15788 | T1179 | NYGTNPGTPPASTSP | |
| NCoA2 | Q15596 | S736 | TIKQEPVSPKKKENA | |
| NDEL1 | Q9GZM8 | T219 | ASLSLPATPVGKGTE | |
| NDEL1 | Q9GZM8 | T245 | GFGTSPLTPSARISA | |
| NeuroD | Q13562 | S162 | EILRSGKSPDLVSFV | |
| NeuroD | Q13562 | S259 | ALEPFFESPLTDCTS | |
| NeuroD | Q13562 | S266 | SPLTDCTSPSFDGPL | |
| NeuroD | Q13562 | S274 | PSFDGPLSPPLSING | |
| NFAT3 | Q14934 | S676 | SNGRRKRSPTQSFRF | + |
| NFL (Neurofilament L) | P07196 | T21 | YKRRYVETPRVHISS | |
| NR0B2 | Q15466 | S26 | AILYALLSSSLKAVP | |
| NR2C1 | P13056 | T222 | SPLTATPTFVTDSES | + |
| NR4A2 | P43354 | S126 | SVYYKPSSPPTPTTP | + |
| NR4A2 | P43354 | T129 | YKPSSPPTPTTPGFQ | + |
| NR4A2 | P43354 | T132 | SSPPTPTTPGFQVQH | + |
| NR4A2 | P43354 | T185 | FKQSPPGTPVSSCQM | |
| Nur77 | P22736 | S140 | GSPCSAPSPSTPSFQ | |
| p27Kip1 | P46527 | S10 | NVRVSNGSPSLERMD | - |
| p27Kip1 | P46527 | S178 | EENVSDGSPNAGSVE | |
| p27Kip1 | P46527 | T187 | NAGSVEQTPKKPGLR | + |
| p47phox | P14598 | S345 | QARPGPQSPGSPLEE | + |
| p53 | P04637 | S15 | PSVEPPLSQETFSDL | + |
| p53 | P04637 | T55 | DDIEQWFTEDPGPDE | + |
| p67phox | P19878 | T233 | EPPPRPKTPEIFRAL | |
| p70S6K (RPS6KB1) | P23443 | S434 | SFEPKIRSPRRFIGS | + |
| p70S6K (RPS6KB1) | P23443 | S447 | GSPRTPVSPVKFSPG | + |
| p70S6K (RPS6KB1) | P23443 | T444 | RFIGSPRTPVSPVKF | + |
| PAK1 | Q13153 | T212 | VIEPLPVTPTRDVAT | - |
| PAPa | P51003 | S537 | DNSMSVPSPTSATKT | |
| PARP1 | P09874 | S372 | VAATPPPSTASAPAA | + |
| PARP1 | P09874 | T373 | AATPPPSTASAPAAV | + |
| PCYT1A | P49585 | S315 | GRMLQAISPKQSPSS | - |
| PDE4B | Q07343 | S659 | YQSMIPQSPSPPLDE | - |
| PDE4C | Q08493 | S641 | YQSKIPRSPSDLTNP | - |
| PDE4D iso1 | Q08499 | S715 | YQSTIPQSPSPAPDD | |
| PDX1 | P52945 | S61 | LGALEQGSPPDISPY | + |
| PDX1 | P52945 | S66 | QGSPPDISPYEVPPL | + |
| PITPNM1 | O00562 | T1223 | AEREGPGTPPTTLAR | |
| PITPNM1 | O00562 | T794 | LEMLVPSTPTSTSGA | ? |
| PKR (PRKR; EIF2AK2) | P19525 | T451 | KRTRSKGTLRYMSPE | - |
| PLCB1 | Q9NQ66 | S982 | KKKSEPSSPDHGSST | |
| PML | P29590 | S36 | PSEGRQPSPSPSPTE | + |
| PML | P29590 | S38 | EGRQPSPSPSPTERA | + |
| PML | P29590 | S40 | RQPSPSPSPTERAPA | |
| PML | P29590 | S527 | PHLDGPPSPRSPVIG | |
| PML | P29590 | S530 | DGPPSPRSPVIGSEV | |
| PML | P29590 | T28 | PTMPPPETPSEGRQP | |
| PPAR-binding protein | Q15648 | T1032 | SSSNRPFTPPTSTGG | |
| PPAR-binding protein | Q15648 | T1457 | HSKSPAYTPQNLDSE | |
| PPARg-1 | P37231 | S112 | AIKVEPASPPYYSEK | - |
| PPP1R2 | P41236 | T72 | MKIDEPSTPYHSMMG | |
| PR | P06401 | S294 | APMAPGRSPLATTVM | ? |
| PTPRR | Q15256 | T361 | EPFVSIPTPREKVAM | |
| PTPRR | Q15256 | Y370 | REKVAMEYLQSASRI | |
| PTPRR | Q15256 | Y44 | SGKPVFIYKHSQDIE | |
| PTPRR | Q15256 | Y62 | DIAPQKIYRHSYHSS | |
| PXN | P49023 | S130 | KQKSAEPSPTVMSTS | |
| PXN | P49023 | S83 | FIHQQPQSSSPVYGS | |
| RIMS1 | Q86UR5 | S447 | ARVSPPESPRARAAA | |
| RIMS1 | Q86UR5 | S731 | ISVISPTSPGALKDA | |
| RORA | P35398 | T216 | PGEAEPLTPTYNISA | - |
| RPS3 | P23396 | T42 | SGVEVRVTPTRTEII | |
| RSK1 (RPS6KA1) | Q15418 | S363 | TSRTPKDSPGIPPSA | + |
| RSK1 (RPS6KA1) | Q15418 | T359 | DTEFTSRTPKDSPGI | + |
| RSK1 (RPS6KA1) | Q15418 | T573 | AENGLLMTPCYTANF | + |
| RSK2 (RPS6KA3) | P51812 | T577 | AENGLLMTPCYTANF | + |
| RXRa | P19793 | S260 | NMGLNPSSPNDPVTN | - |
| Sam68 | Q07666 | S58 | SRGGARASPATQPPP | |
| Sam68 | Q07666 | T84 | TVGGPAPTPLLPPSA | |
| Securin | O95997 | S165 | LFQLGPPSPVKMPSP | |
| Separase | Q14674 | S1126 | IAPSTNSSPVLKTKP | |
| SH2-Bb | Q9NRF2 | S96 | PPILAPLSPGAEISP | |
| Shc1 | P29353 | S36 | TPPEELPSPSASSLG | - |
| SLC9A1 (NHE1) | P19634 | S770 | MMRSKETSSPGTDDV | + |
| Smad1 | Q15797 | S187 | NSHPFPHSPNSSYPN | |
| Smad1 | Q15797 | S195 | PNSSYPNSPGSSSST | |
| Smad1 | Q15797 | S206 | SSSTYPHSPTSSDPG | |
| Smad1 | Q15797 | S214 | PTSSDPGSPFQMPAD | |
| Smad2 | Q15796 | S245 | NQSMDTGSPAELSPT | |
| Smad2 | Q15796 | S250 | TGSPAELSPTTLSPV | |
| Smad2 | Q15796 | S255 | ELSPTTLSPVNHSLD | |
| Smad2 | Q15796 | T220 | QSNYIPETPPPGYIS | |
| Smad3 | P84022 | S204 | NHSMDAGSPNLSPNP | |
| Smad3 | P84022 | S208 | DAGSPNLSPNPMSPA | |
| Smad3 | P84022 | S213 | NLSPNPMSPAHNNLD | - |
| Smad3 | P84022 | T179 | PQSNIPETPPPGYLS | - |
| Smad4 | Q13485 | T277 | GSRTAPYTPNLPHHQ | |
| SOS1 | Q07889 | S1132 | TLPHGPRSASVSSIS | |
| SOS1 | Q07889 | S1167 | ESAPAESSPSKIMSK | |
| SOS1 | Q07889 | S1178 | IMSKHLDSPPAIPPR | |
| SOS1 | Q07889 | S1193 | QPTSKAYSPRYSISD | |
| SOS1 | Q07889 | S1197 | KAYSPRYSISDRTSI | |
| SP1 | P08047 | S59 | GGQESQPSPLALLAA | |
| SP1 | P08047 | T453 | SGPIIIRTPTVGPNG | |
| SP1 | P08047 | T739 | SEGSGTATPSALITT | |
| SP3 | Q02447 | S73 | CSKIGPPSPGDDEEE | ? |
| SPHK1 | Q9NYA1 | S311 | VGSKTPASPVVVQQG | + |
| SPHK2 | Q9NRA0 | S387 | PATVEPASPTPAHSL | |
| SPHK2 | Q9NRA0 | T614 | AFRLEPLTPRGVLTV | |
| Spinophilin | Q96SB3 | S15 | GPGGPLRSASPHRSA | |
| Spinophilin | Q96SB3 | S203 | KLDADAVSPTVSQLS | |
| SREBP-2 | Q12772 | S432 | NQNVLLMSPPASDSG | |
| SREBP-2 | Q12772 | S455 | SIDSEPGSPLLDDAK | |
| STAT1 | P42224 | S727 | TDNLLPMSPEEFDEM | - |
| STAT3 | P40763 | S727 | NTIDLPMSPRTLDSL | - |
| STAT5A | P42229 | S726 | TYMDQAPSPAVCPQA | |
| STAT5A | P42229 | S780 | DSLDSRLSPPAGLFT | - |
| STF-1 | Q13285 | S203 | EYPEPYASPPQPGLP | |
| STMN2 | Q93045 | S62 | EVLQKAPSPISEAPR | |
| STMN2 | Q93045 | S73 | EAPRTLASPKKKDLS | |
| supervillin (ferret) | O95425 | Y132 | DPEADSEYLSRYTKS | |
| SYN1 | P17600 | S551 | PAARPPASPSPQRQA | |
| SYN1 | P17600 | S62 | SGVAPAASPAAPSPG | |
| SYN1 | P17600 | S67 | AASPAAPSPGSSGGG | |
| SYN3 | O14994 | S470 | PQGQQPLSPQSGSPQ | |
| Tau iso5 (Tau-C) | P10636-5 | S202 | SGYSSPGSPGTPGSR | |
| Tau iso5 (Tau-C) | P10636-5 | T205 | SSPGSPGTPGSRSRT | |
| Tau iso8 | P10636-8 | T181 | KTPPAPKTPPSSGEP | |
| Tau iso9 (Tau-F) | P10636-9 | S404 | PVVSGDTSPRHLSNV | |
| TH | P07101 | S61 | SYTPTPRSPRFIGRR | |
| TH | P07101 | S62 | SYTPTPRSPRFIGRR | |
| TH | P07101 | T8 | MPTPDATTPQAKGFR | |
| THRB | P10828 | S142 | IQKNLHPSYSCKYEG | + |
| TNF-R1 | P19438 | S274 | LAPNPSFSPTPGFTP | |
| TNF-R1 | P19438 | S286 | FTPTLGFSPVPSSTF | |
| TNF-R1 | P19438 | T276 | PNPSFSPTPGFTPTL | |
| TNF-R1 | P19438 | T280 | FSPTPGFTPTLGFSP | |
| TNF-R1 | P19438 | T300 | FTSSSTYTPGDCPNF | |
| TOB1 | P50616 | S152 | PASSVSSSPSPPFGH | |
| TOB1 | P50616 | S154 | SSVSSSPSPPFGHSA | |
| TOB1 | P50616 | S164 | FGHSAAVSPTFMPRS | |
| TOP2A | P11388 | S1213 | QMAEVLPSPRGQRVI | |
| TOP2A | P11388 | S1247 | KNENTEGSPQEDGVE | |
| TOP2A | P11388 | S1354 | DFVPSDASPPKTKTS | |
| TOP2A | P11388 | S1361 | SPPKTKTSPKLSNKE | |
| TOP2A | P11388 | S1393 | GSVPLSSSPPATHFP | |
| TPPP | O94811 | S160 | GVTKAISSPTVSRLT | |
| TPPP | O94811 | S18 | ANRTPPKSPGDPSKD | |
| TSC2 | P49815 | S664 | KKTSGPLSPPTGPPG | |
| TTK | P33981 | S821 | GQLVGLNSPNSILKA | ? |
| UBF | P17480 | T117 | DFPKKPLTPYFRFFM | - |
| UBF | P17480 | T201 | DIPEKPKTPQQLWYT | - |
| Vinexin | O60504 | S530 | DGPQLPTSPRLTAAA |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
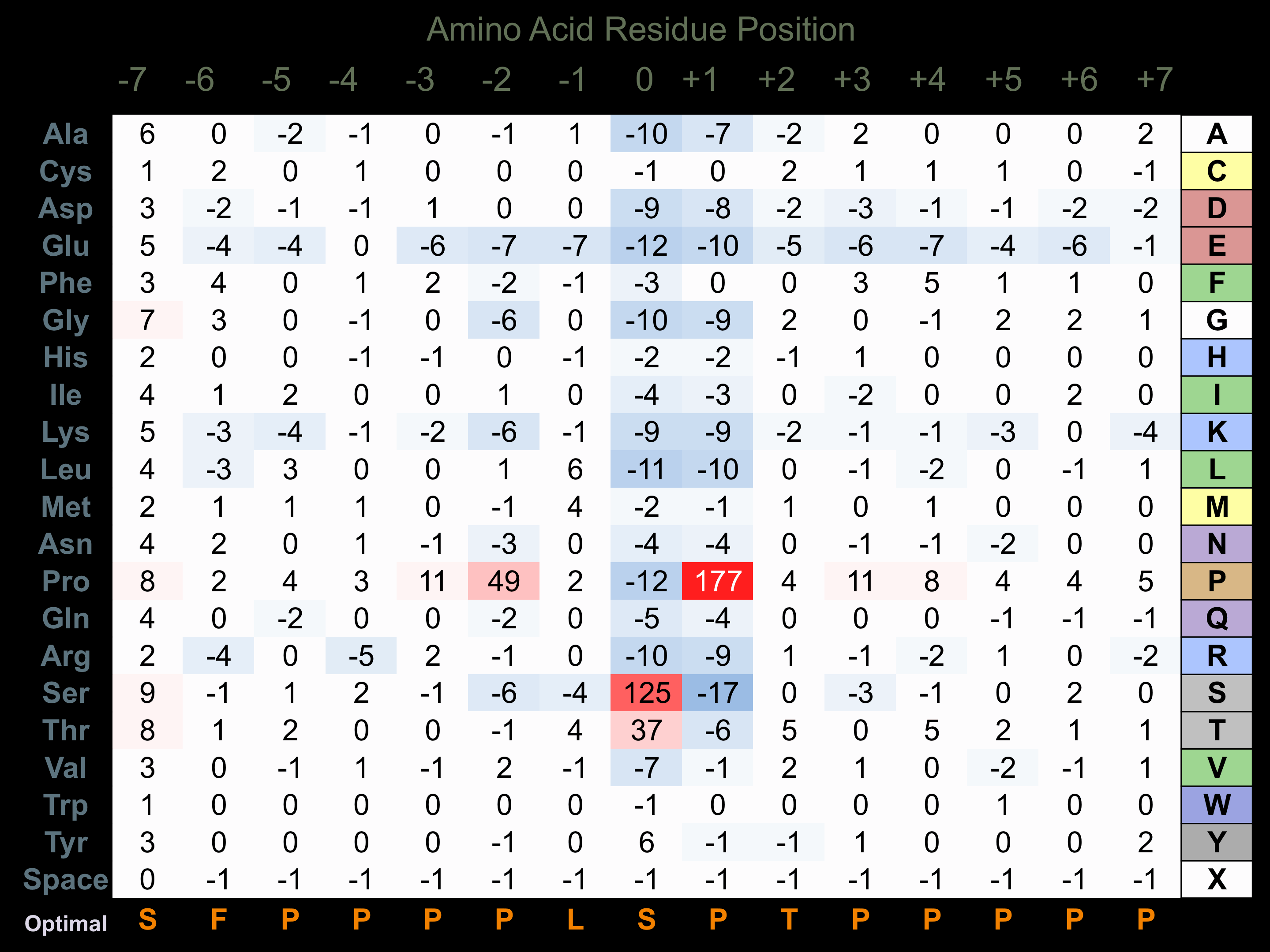
Matrix Type:
Experimentally derived from alignment of 474 known protein substrate phosphosites and 90 peptides phosphorylated by recombinant ERK2 in vitro tested in-house by Kinexus. Note that additional binding sites on ERK2 substrates with D motifs (consensus=L-x-e-R/k-R/k-x-x-x-x-x-l/i/v-p-p or p-x-L/v/i-x-p-p-x-x-x-x-l-l-x-r/k-k/r-R/k-K/r) and/or FxFP motifs (Phe-any amino acid-Phe-Pro) facilitate higher selectivity for phosphorylation by this protein kinase.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, eye, kidney, skin, and neurological disorders
Specific Diseases (Non-cancerous):
Trachoma; Corticobasal degeneration (CBGD); Congenital insensitivity to pain with anhidrosis (CIPA); Mesangial proliferative glomerulonephritis; Frozen shoulder; Retrograde amnesia
Comments:
Trachoma is a rare infectious disease that affects the eye. Transmission is through direct contact with infected eye discharge. Corticobasal Degeneration (CBGD) is a rare neuronal disease characterized by neuronal loss and reduced brain size. Symptoms may be similar to Parkinson disease. CBGD can affect the cortex, whole brain, and eye tissues. The rare disorder Congenital Insensitivity to Pain with Anhidrosis (CIPA) is characterized by pain insensitivity and reduced or absent sweating. CIPA sufferers may have repeated bone infections and affected tissues can include bone and skin. Mesangial Proliferative Glomerulonephritis is a rare disease and is a form of Lupus Nephritis, which is the inflammation of the kidney. Anhidrosis is the decrease or fully impaired ability to sweat, leading to fever-related issues. Frozen Shoulder is characterized by inflammation of the shoulder, causing pain, and it is associated with diabetes mellitus and osteoarthritis. Frozen Shoulder can affect bone, skin, and testes. The K54R mutation does not inhibit MAP2K1-ERK2 interaction. The T185A and Y187A mutations, in conjunction, will inhibit ERK2 interaction with TPR, whereas L234A can inhibit a TPR complex by itself. PTPRJ-mediated phosphorylation of ERK2 is lost with a D318A mutation. MEK2 interaction with MAP2K1, but not TPR, is inhibited with the D318N+D321N mutations.
Specific Cancer Types:
Breast cancer; Pancreatic cancer; Thyroid lymphomas; Ewing's Family of tumours; Distal 22q11.2 Microdeletion syndrome
Comments:
ERK2 appears to be a tumour requiring protein (TRP). The active form of the protein kinase normally acts to promote tumour cell proliferation. ERK2 has been linked with the Ewing's Family of tumours, which are rare cancers that are similar to medullablastoma and cerebral primitive neuroectodermal tumours. Distal 22q11. 2 Microdeletion Syndrome is a rare fetal cancer disease, which is similar to B-cell chronic lymphocytic leukemia and myeloid leukemia.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +65, p<0.002); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +223, p<0.088); Gastric cancer (%CFC= +74, p<0.008); Prostate cancer (%CFC= -54, p<0.016); and Uterine fibroids (%CFC= +62, p<0.025). The COSMIC website notes an up-regulated expression score for ERK2 in diverse human cancers of 439, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 355 for this protein kinase in human cancers was 5.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25487 diverse cancer specimens. This rate is only 22 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.41 % in 805 skin cancers tested; 0.3 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: E332K (12).
Comments:
Only 2 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.