Nomenclature
Short Name:
MEK2
Full Name:
Dual specificity mitogen-activated protein kinase kinase 2
Alias:
- EC 2.7.12.2
- ERK activator kinase 2
- MKK2
- MP2K2
- PRKMK2EC
- MEK2
- MAP kinase kinase 2
- MAP2K2
- MAPK,ERK kinase 2
- MAPKK2
Classification
Type:
Dual specificity protein kinase
Group:
STE
Family:
STE7
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
44,424
# Amino Acids:
400
# mRNA Isoforms:
1
mRNA Isoforms:
44,424 Da (400 AA; P36507)
4D Structure:
Interacts with MORG1
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 26 | 58 | Coiled-coil |
| 72 | 327 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S23, S26, S94, S198, S216-, S222+, S226+, S293, S295, S306, S312, S372.
Acetylated:
T13, S222, S226.
Threonine phosphorylated:
T13, T59, T230-, T394, T396, T398.
Ubiquitinated:
K40, K51, K61, K63, K68, K101, K108, K196, K209.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 35
35
651
42
841
 9
9
169
20
144
 5
5
100
3
97
 29
29
533
130
607
 41
41
768
35
787
 5
5
88
115
114
 22
22
415
47
579
 51
51
948
50
1301
 35
35
660
24
572
 9
9
159
116
153
 8
8
154
34
167
 38
38
715
246
646
 6
6
119
36
139
 7
7
124
15
123
 8
8
144
25
146
 15
15
283
22
325
 5
5
99
284
103
 9
9
163
14
176
 19
19
347
131
341
 27
27
511
162
603
 10
10
185
22
201
 11
11
210
28
276
 20
20
371
14
313
 18
18
332
14
333
 9
9
166
22
174
 100
100
1861
73
3831
 7
7
138
39
153
 7
7
121
14
111
 7
7
124
14
118
 39
39
720
42
722
 33
33
618
30
572
 31
31
574
51
549
 15
15
281
90
459
 39
39
719
83
584
 51
51
953
45
2766
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 79.5
79.5
87.5
93 70.9
70.9
72.5
93 -
-
-
97 -
-
-
89 -
-
-
97 -
-
-
- 93.8
93.8
97.3
94 94
94
98
94 -
-
-
- 64.5
64.5
73
- 93.5
93.5
96.3
94 79
79
87.5
92 32
32
50.8
86 -
-
-
- 62.3
62.3
78.3
- -
-
-
- 51.8
51.8
66.5
- 63.6
63.6
75.7
- -
-
-
- -
-
-
- -
-
-
44 32.8
32.8
50.3
43 -
-
-
45 -
-
-
43
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAPK3 - P27361 |
| 2 | RAF1 - P04049 |
| 3 | MAPK1 - P28482 |
| 4 | BRAF - P15056 |
| 5 | KSR1 - Q8IVT5 |
| 6 | ARAF - P10398 |
| 7 | CNKSR1 - Q969H4 |
| 8 | CASP9 - P55211 |
| 9 | MAPKSP1 - Q9UHA4 |
| 10 | DUSP3 - P51452 |
| 11 | GRIN1 - Q05586 |
| 12 | GRIN2D - O15399 |
| 13 | COPS5 - Q92905 |
| 14 | RGS12 - O14924 |
| 15 | MAPK14 - Q16539 |
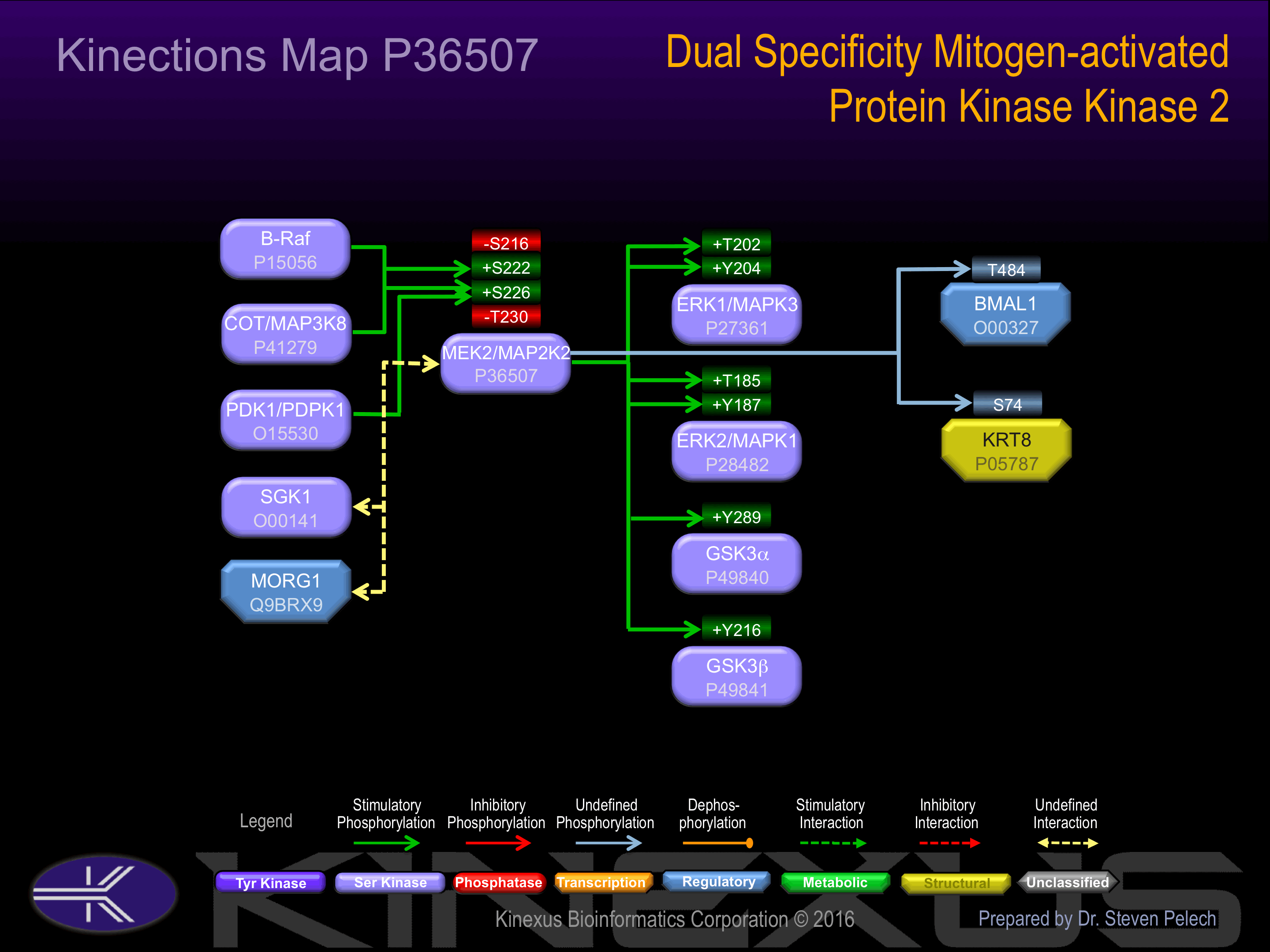
Regulation
Activation:
Phosphorylation of Ser-222 and Ser226 increases phosphotransferase activity.
Inhibition:
Phosphorylation of Ser-216 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| BMAL1 | O00327 | T484 | SPLNITSTPPPDASS | |
| ERK1 (MAPK3) | P27361 | T202 | HDHTGFLTEYVATRW | + |
| ERK1 (MAPK3) | P27361 | Y204 | HTGFLTEYVATRWYR | + |
| ERK2 (MAPK1) | P28482 | T185 | HDHTGFLTEYVATRW | + |
| ERK2 (MAPK1) | P28482 | Y187 | HTGFLTEYVATRWYR | + |
| GSK3a | P49840 | Y279 | RGEPNVSYICSRYYR | + |
| GSK3b | P49841 | Y216 | RGEPNVSYICSRYYR | + |
| KRT8 | P05787 | S74 | TVNQSLLSPLVLEVD |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
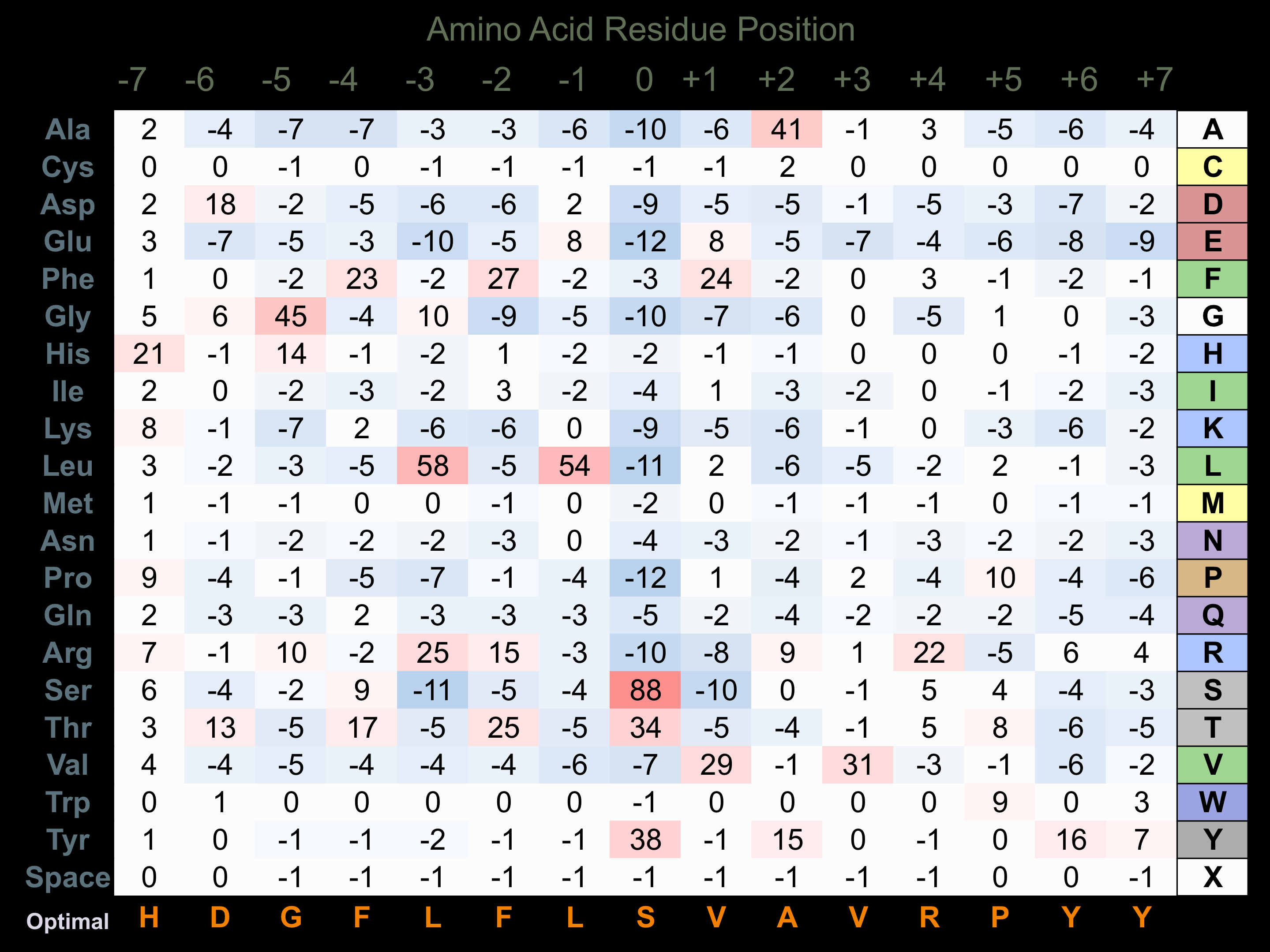
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Skin, heart, and neuronal disorders
Specific Diseases (Non-cancerous):
Cardiofaciocutaneous syndrome; Noonan syndrome; Costello syndrome; Cardiofaciocutaneous syndrome 4; Leopard syndrome; Neurofibromatosis-Noonan syndrome (NFNS); Map2k2-related cardiofaciocutaneous syndrome
Comments:
The rare Cardiofaciocutaneous Syndrome can affect the heart (cardio-), face (facio-), skin, and hair (cutaneous). Symptoms can include mild to severe cognitive disability, and developmental delay. Noonan Syndrome is a rare disorder that involves the thickening and hardening of the heart muscles. Costello Syndrome sufferers share many of the same symptoms as Cardiofaciocutaneous Syndrome, including mild to severe cognitive disability, and developmental delay, but they also have characteristic facial features, extremely flexible joints, folds of loose skin, and an unusually fast heartbeat. In Cardiofaciocutaneous Syndrome 4 (CFC4) a mutation in MEK2, P128Q, can lead to an increase in kinase phosphotransferase activity. Leopard Syndrome is a rare yet very distinctive disorder where sufferers are marked with spots on their skin. They also suffer from abnormalities in the heart, widely spaced eyes, abnormal genitalia, short stature, and hearing loss. Neurofibromatosis-Noonan Syndrome (NFNS) is a rare disorder characterized by cognitive impairment, psychomotor impairment, learning disabilities, dwarfism, and abnormal development of the torso.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +106, p<0.062); Brain oligodendrogliomas (%CFC= +209, p<0.002); Large B-cell lymphomas (%CFC= +53, p<0.006); Skin melanomas - malignant (%CFC= +274, p<0.0001); and Uterine leiomyomas (%CFC= +47, p<0.082). The COSMIC website notes an up-regulated expression score for MEK2 in diverse human cancers of 406, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 30 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 26285 diverse cancer specimens. This rate is only -3 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.31 % in 1280 large intestine cancers tested; 0.31 % in 1139 skin cancers tested.
Frequency of Mutated Sites:
None > 4 in 21,282 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.