Nomenclature
Short Name:
FLT3
Full Name:
Fetal liver fms-related tyrosine kinase 3
Alias:
- CD135
- FLT-3
- Fms-related tyrosine kinase 3
- Kinase Flt3
- STK-1
- EC 2.7.10.1
- Fetal liver kinase 2
- FL cytokine receptor
- FLK2
- FLK-2
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
PDGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
112903
# Amino Acids:
993
# mRNA Isoforms:
2
mRNA Isoforms:
112,903 Da (993 AA; P36888); 108,378 Da (952 AA; P36888-2)
4D Structure:
Interacts with FIZ1 following ligand activation
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
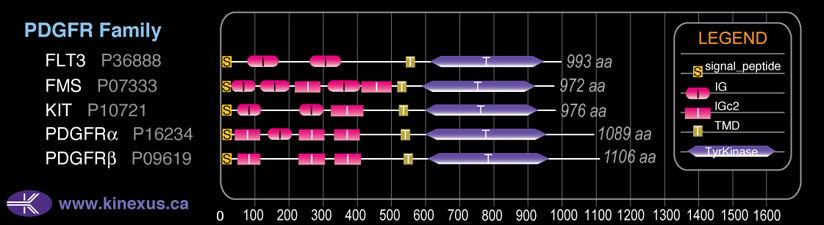
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N43, N100, N151, N306, N323, N351, N354, N473, N502, N541.
Serine phosphorylated:
S759, S840, S993.
Threonine phosphorylated:
T343, T728, T968.
Tyrosine phosphorylated:
Y166, Y416, Y572, Y589-, Y591+, Y597, Y599+, Y630, Y726, Y768+, Y793, Y842-, Y889+, Y919+, Y955, Y969+.
Ubiquitinated:
K602, K614, K623, K634, K644, K719, K772, K932.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1013
16
1338
 3
3
28
11
21
 2
2
23
24
38
 17
17
168
71
348
 32
32
324
14
323
 1
1
15
37
8
 41
41
412
23
579
 41
41
419
45
1212
 21
21
213
10
201
 12
12
118
77
159
 3
3
32
41
44
 31
31
316
151
486
 6
6
60
35
32
 3
3
32
12
29
 6
6
63
37
56
 2
2
17
9
22
 2
2
22
203
18
 3
3
32
33
31
 4
4
43
70
41
 25
25
258
56
314
 5
5
53
37
40
 4
4
42
38
32
 1
1
14
30
17
 3
3
32
31
51
 5
5
55
37
45
 93
93
947
50
2682
 3
3
34
38
26
 3
3
28
33
41
 2
2
17
30
22
 17
17
169
14
123
 45
45
460
18
328
 59
59
598
21
638
 8
8
79
52
193
 68
68
691
31
720
 13
13
130
22
82
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99
99
99.5
99 99
99
99.5
96.5 -
-
-
91 -
-
-
89 96.5
96.5
98.5
92 30.3
30.3
48.5
- -
-
-
86 84
84
89.7
84.5 29.6
29.6
47.6
- 66.9
66.9
80.6
- 66.9
66.9
80.6
57 31.5
31.5
48.4
52 30.7
30.7
46.6
46 33.8
33.8
49.8
- 30.6
30.6
47.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
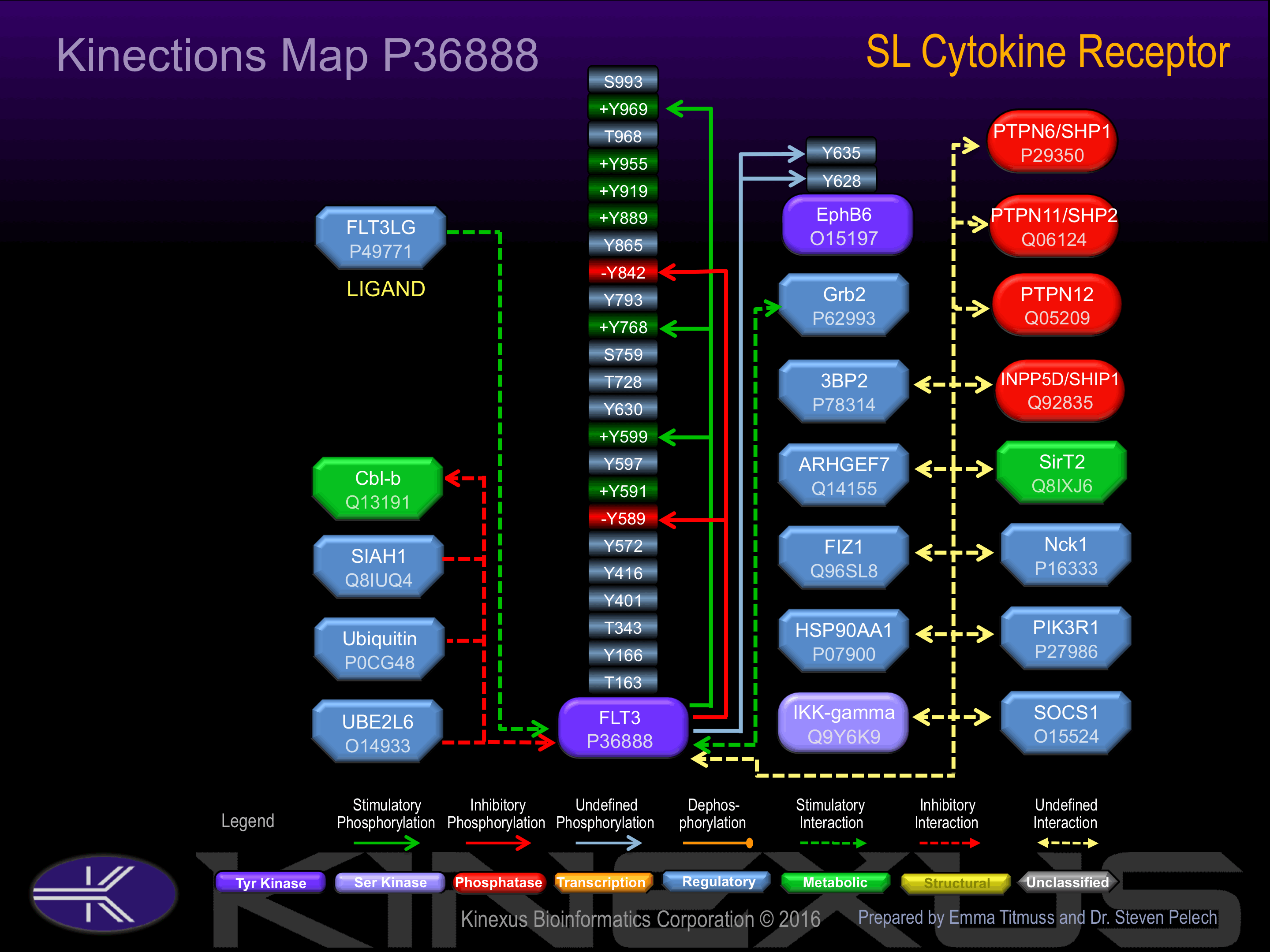
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PTPN6 - P29350 |
| 2 | PTPN12 - Q05209 |
| 3 | FLT3LG - P49771 |
| 4 | CBLB - Q13191 |
| 5 | NCK1 - P16333 |
| 6 | FIZ1 - Q96SL8 |
| 7 | PIK3R1 - P27986 |
Regulation
Activation:
Phosphorylation of Tyr-591 increases phosphotransferase activity. Phosphorylation of Tyr-599 increases phosphotransferase activity and induces interaction with SHP2 and Src.
Inhibition:
Phosphorylation of Tyr-589 inhibits phosphotransferase activity and induces interaction with Src. Phosphorylation of Tyr-842 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| EphB6 | O15197 | Y628 | RKRRGTGYTEQLQQY | |
| EphB6 | O15197 | Y635 | YTEQLQQYSSPGLGV | |
| FLT3 | P36888 | Y589 | TGSSDNEYFYVDFRE | - |
| FLT3 | P36888 | Y591 | SSDNEYFYVDFREYE | + |
| FLT3 | P36888 | Y599 | VDFREYEYDLKWEFP | + |
| FLT3 | P36888 | Y726 | KEHNFSFYPTFQSHP | |
| FLT3 | P36888 | Y842 | DIMSDSNYVVRGNAR | - |
| FLT3 | P36888 | Y955 | ADAEEAMYQNVDGRV | ? |
| FLT3 | P36888 | Y969 | VSECPHTYQNRRPFS | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
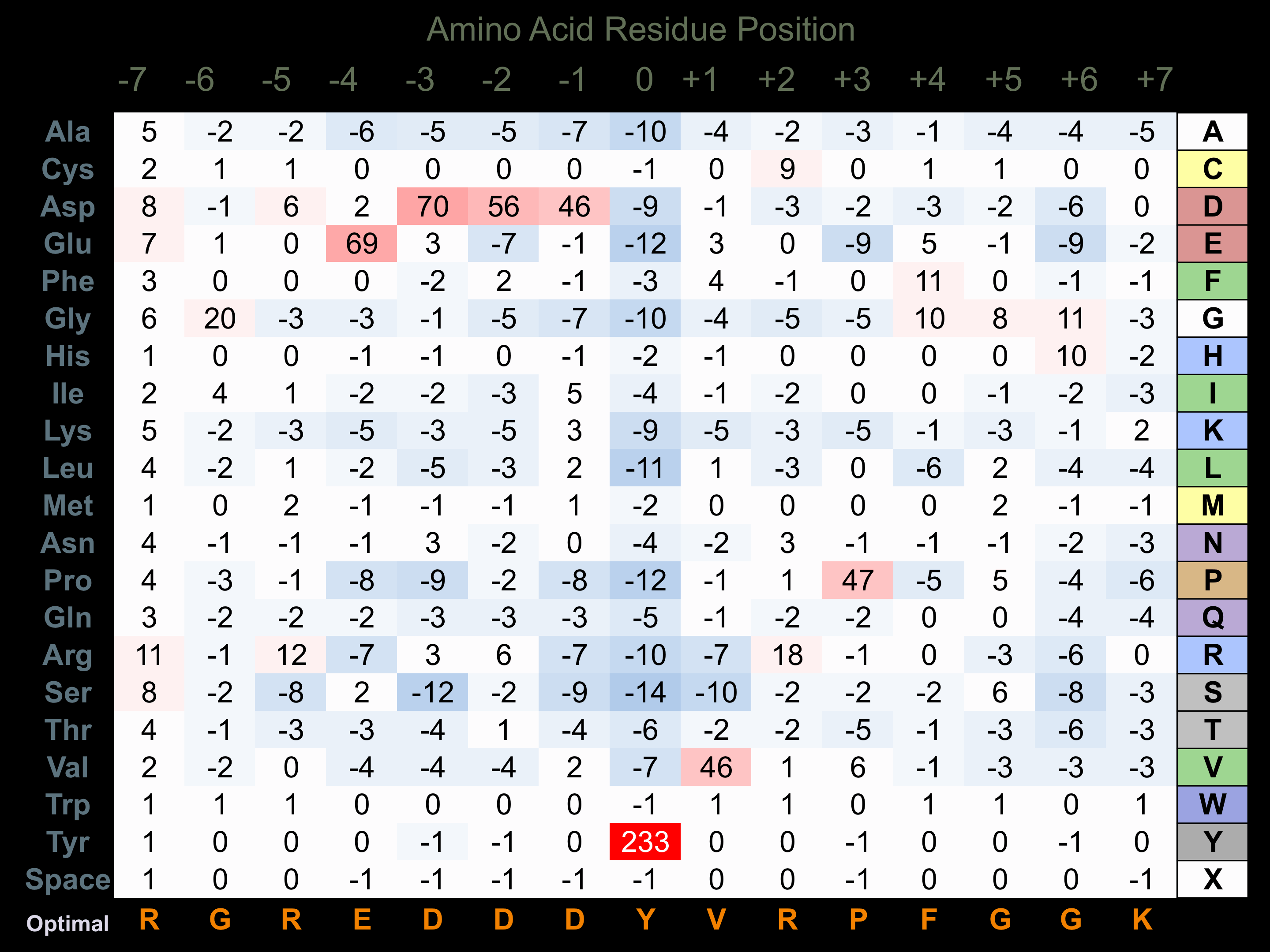
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Acute myelogenous leukemias (AML); Myeloid leukemias; Leukemias; Lymphoblastic leukemias; Juvenile myelomonocytic leukemias (JMML); Gastrointestinal stromal tumours (GANT); Myelodysplastic syndromes (MSD); Acute promyelocytic leukemias (APL); Cytogenetically normal Acute myeloid leukemias (AML); Core binding factor Acute myeloid leukemias (AML); Chronic myelomonocytic leukemias (CMML); Tetraploidy; Myeloid sarcomas; Acute myeloid leukemias (AML) with T(6;9)(p23;q34); Chronic myeloproliferative disease (CMPD); 8p11 Myeloproliferative syndrome; Bone marrow cancer; Acute myelomonocytic leukemias (AMML); Acute biphenotypic leukemias (BAL); Precursor T-cell acute lymphoblastic leukemias; Leukemias, acute lymphoblastic 3 (B-ALL); Acute myeloblastic leukemias with maturation; Acute myeloid leukemias (AML) with T(8;21)(q22;q22) Translocation; Acute myeloid leukemias (AML) with abnormal bone marrow eosinophils Inv(16)(p13q22) or T(16;16)(p13;q22); Acute myeloblastic leukemias (AML) without maturation; Leukemias, acute lymphoblastic, somatic; Minimally differentiated acute myeloblastic leukemias (AML)
Comments:
FLT3 is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. FLT3 is linked with Acute Myeloid Leukemia (AML), which is a rare blood cancer disease which will displace healthy blood cells and can lead to fever, shortness of breath, fatigue, and easy bruising. Myeloid Leukemia is a cancer of the myeloid progenitor cells and it can affect myeloid, bone, and bone marrow. Lymphoblastic Leukemia is related to leukemia and can affect T cells, B cells, and bone tissue. Juvenile Myelomonocytic Leukemia (JMML) is a rare chronic blood leukemia that mostly affects young (under 4 years) children. Myelodysplastic Syndromes (MSD) is a rare bone marrow cancer resulting in insufficient functioning and circulating red blood cells. Acute Promyelocytic Leukemia is also a rare bone marrow cancer. Tetraploidy is a rare chromosomal aberration disorder that is related to leukemia. The rare cancer Myeloid Sarcoma is characterized by an accumulation of white blood cells forming a solid cellular mass. Chronic Myeloproliferative Disease is another rare cancer of the bone marrow which results in impaired red blood cell production. Chronic Myeloproliferative Disease (CMPD) is a slow growing cancer. Acute Biphenotypic Leukemia (BAL) is a rare cancer arising from both the lymphoid and myeloid lineages. BAL is related to the complement cascade and clotting cascade pathways. Precursor T-Cell Acute Lymphoblastic Leukemia is a rare cancer and some of its related pathways include the gefitinib pathway, and the cytokine inflammatory response. Leukemia, Acute Lymphoblastic 3 (B-ALL) is a rare cancer which is related to the gefitinib and the transcription factors in neurogenesis pathways. Acute Myeloblastic Leukemia with Maturation is a sub-type of AML. Acute Myeloblastic Leukemia Without Maturation is a rapidly progressing form of leukemia where white blood cells are overproduced in the bone marrow. Minimally Differentiated Acute Myeloblastic Leukemia is a subtype of AML. Gastrointestinal Stromal tumour (GANT) is a soft tissue sarcoma growing from the stomach, intestines, or rectum. STAT5A activation can be inhibited with a Y589F or Y591F mutation. The Y599F mutation can interrupt FLT3 complex formation with PTPN11/SHP2. FLT3 Kinase phosphotransferase activity can abolished with a K644A mutation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +357, p<0.057); and Breast epithelial carcinomas (%CFC= +147, p<0.037). The COSMIC website notes an up-regulated expression score for FLT3 in diverse human cancers of 299, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 1.53 % in 103570 diverse cancer specimens. This rate is 20.45-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 2.06 % in 74463 haematopoietic and lymphoid cancers tested; 0.4 % in 1468 skin cancers tested; 0.38 % in 1712 large intestine cancers tested; 0.15 % in 743 endometrium cancers tested; 0.13 % in 856 stomach cancers tested; 0.13 % in 2889 lung cancers tested; 0.12 % in 1270 liver cancers tested; 0.11 % in 1176 ovary cancers tested; 0.05 % in 2001 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: D835Y (211); D835? (707); D835E (27).
Comments:
A large number of complex mutations and insertions mutations are clustered between amino acid residues 569 and 627. Over 50 deletionsmutations occurred at amino acid residues 835 and 836. These mutation patterns are consistent with FLT3 as an oncoprotein.