Nomenclature
Short Name:
ICK
Full Name:
Serine-threonine-protein kinase ICK
Alias:
- HICK
- Laryngeal cancer kinase 2
- LCK2
- MAK-related kinase
- MGC46090
- Serine/threonine kinase ICK; ECO; MRK
- Intestinal cell (MAK-like) kinase
- Intestinal cell kinase
- KIAA0936
- Kinase ICK
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
RCK
SubFamily:
NA
Structure
Mol. Mass (Da):
71,427
# Amino Acids:
632
# mRNA Isoforms:
2
mRNA Isoforms:
71,427 Da (632 AA; Q9UPZ9); 34,071 Da (292 AA; Q9UPZ9-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
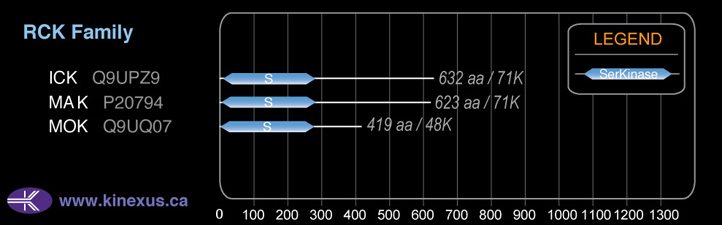
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 4 | 284 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K562.
Serine phosphorylated:
S17, S23, S53, S254, S528, S529, S585.
Threonine phosphorylated:
T6, T14, T157+, T248, T463.
Tyrosine phosphorylated:
Y15, Y156+, Y159+, Y177, Y583.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 24
24
832
22
910
 1.3
1.3
44
11
48
 11
11
388
27
313
 6
6
209
94
346
 16
16
565
21
472
 0.4
0.4
14
46
18
 6
6
199
29
451
 16
16
542
44
1762
 12
12
414
10
416
 1.5
1.5
51
69
70
 2
2
81
38
92
 11
11
381
126
374
 4
4
140
37
160
 0.9
0.9
31
9
16
 3
3
107
36
127
 3
3
106
13
140
 8
8
265
132
2016
 13
13
441
33
415
 2
2
76
69
95
 17
17
606
79
629
 8
8
290
36
277
 5
5
171
35
171
 7
7
237
28
183
 6
6
199
34
197
 5
5
163
35
165
 9
9
298
70
298
 7
7
227
41
248
 9
9
307
34
316
 7
7
231
34
228
 0.4
0.4
14
14
21
 28
28
994
30
550
 100
100
3491
26
6554
 4
4
134
56
344
 15
15
533
52
464
 0.9
0.9
30
35
20
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99
99
99.5
99 86.8
86.8
90
95 -
-
-
92 -
-
-
- 86.1
86.1
89.4
92 -
-
-
- 86.3
86.3
92
87 85.5
85.5
91.6
86 -
-
-
- 56.6
56.6
68.8
- -
-
-
76 23.1
23.1
32.1
62 47
47
60.5
- -
-
-
- -
-
-
- 45.8
45.8
61.3
- -
-
-
67 48.5
48.5
62.1
- -
-
-
- -
-
-
- -
-
-
57 34.9
34.9
48.8
56 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
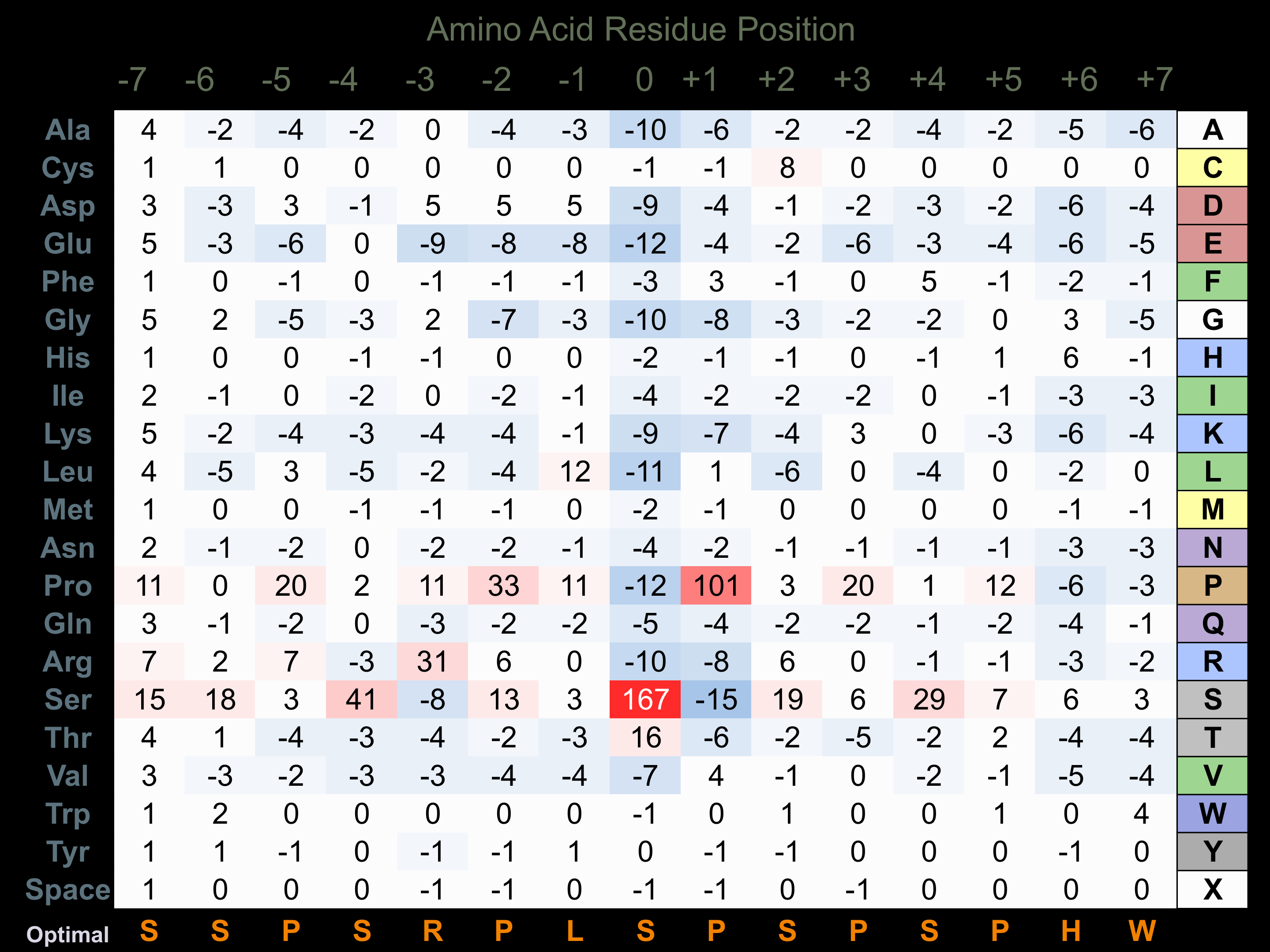
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Alvocidib | Kd = 690 pM | 9910986 | 428690 | 22037378 |
| R547 | Kd = 2.2 nM | 6918852 | 22037378 | |
| AT7519 | Kd = 8.3 nM | 11338033 | 22037378 | |
| Staurosporine | Kd = 16 nM | 5279 | 22037378 | |
| Lestaurtinib | Kd = 39 nM | 126565 | 22037378 | |
| AC1NS7CD | Kd = 150 nM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 150 nM | 3025986 | 296468 | 22037378 |
| Ruboxistaurin | Kd = 240 nM | 153999 | 91829 | 22037378 |
| PHA-665752 | Kd = 400 nM | 10461815 | 450786 | 22037378 |
| Sunitinib | Kd = 470 nM | 5329102 | 535 | 19654408 |
| A674563 | Kd = 550 nM | 11314340 | 379218 | 22037378 |
| Enzastaurin | Kd = 680 nM | 176167 | 300138 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| SU14813 | Kd = 1.2 µM | 10138259 | 1721885 | 22037378 |
| Nintedanib | Kd = 1.7 µM | 9809715 | 502835 | 22037378 |
| TG101348 | Kd = 1.9 µM | 16722836 | 1287853 | 22037378 |
| KW2449 | Kd = 2.2 µM | 11427553 | 1908397 | 22037378 |
| AST-487 | Kd = 2.5 µM | 11409972 | 574738 | 22037378 |
| NVP-TAE684 | Kd = 4 µM | 16038120 | 509032 | 22037378 |
Disease Linkage
General Disease Association:
Bone disorder (fetal)
Specific Diseases (Non-cancerous):
Endocrine-cerebroosteodysplasia
Comments:
Endocrine-cerebro-osteodysplasia (ECO) is a neonatal lethal recessive genetic disease that is characterized by multiple abnormalities affecting the endocrine system, nervous system (specifically the cerebrum), and the skeletal system. ECO has been linked to a missense mutation in the ICK gene, however it is not known how a non-functional ICK results in the ECO phenotype or the pathological mechanisms of the ECO phenotype. In animal studies, mice embryos that lack ICK display cleft palate, hydrocephalus, polydactyly, and delayed development of the skeletal system, which closely resembles the ECO phenotype. In cultured cells, knockout of ICK expression results in the elongation of cilia and abnormal sonic hedgehog (Shh) expression, implicating aberrant Shh signalling as potential conrtibutor to the ECO phenotype. In addition, wild-type ICK proteins are usually localized near the basal body in the proximal region of the cilia, compared to the mutant ICK that accumulates in the distal end of the bulged cilia. Consistent with these in vitro observations, mice lacking ICK showed elongated cilia and a reduction in Shh signalling during limb and digit morphogenesis, leading to development defects. Therefore, ICK is proposed to play a critical role in the regulation of ciliary length and loss of this function leads to ciliary defects correlated with abnormal Shh signalling during development, which together may contribute to the pathogenesis of congenital diseases such as ECO.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for ICK in diverse human cancers of 411, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 47 for this protein kinase in human cancers was 0.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24896 diverse cancer specimens. This rate is only -19 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.31 % in 864 skin cancers tested; 0.29 % in 1270 large intestine cancers tested; 0.24 % in 603 endometrium cancers tested; 0.21 % in 589 stomach cancers tested; 0.09 % in 1822 lung cancers tested; 0.08 % in 1316 breast cancers tested; 0.07 % in 1512 liver cancers tested; 0.06 % in 273 cervix cancers tested; 0.04 % in 833 ovary cancers tested; 0.03 % in 548 urinary tract cancers tested; 0.03 % in 1446 kidney cancers tested; 0.02 % in 881 prostate cancers tested; 0.02 % in 710 oesophagus cancers tested; 0.02 % in 2082 central nervous system cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested; 0.01 % in 1459 pancreas cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: A155V (4).
Comments:
Only 5 deletions, 1 complex mutation and no insertions are noted on the COSMIC website.

