Nomenclature
Short Name:
ILK
Full Name:
Integrin-linked protein kinase 1
Alias:
- 59 kDa serine/threonine protein kinase
- Kinase ILK
- P59ILK
- EC 2.7.11.1
- ILK1
- ILK-1
- Integrin-linked kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
MLK
SubFamily:
ILK
Specific Links
Structure
Mol. Mass (Da):
51,419
# Amino Acids:
452
# mRNA Isoforms:
3
mRNA Isoforms:
51,419 Da (452 AA; Q13418); 44,349 Da (391 AA; Q13418-2); 36,467 Da (318 AA; Q13418-3)
4D Structure:
Interacts with cytoplasmic domain of beta 1 subunit of integrin. Could also interacts with beta 2, beta 3 and/or beta 5 subunit of integrin. Interacts (via ANK repeats) with LIMS1 and LIMS2. Interacts with parvins and probably TGFB1I1.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K426.
Serine phosphorylated:
S76, S186, S232, S246, S343+.
Threonine phosphorylated:
T69, T167, T172, T173, T181+.
Tyrosine phosphorylated:
Y351+.
Ubiquitinated:
K85, K139, K165, K170, K191, K198, K209, K220, K438, K448.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 66
66
939
22
670
 30
30
424
10
95
 98
98
1392
13
625
 59
59
836
58
1267
 79
79
1128
14
825
 19
19
272
43
487
 41
41
585
19
563
 88
88
1254
33
1440
 65
65
919
10
550
 31
31
440
57
244
 24
24
346
26
242
 77
77
1099
117
760
 27
27
377
24
293
 31
31
442
9
190
 26
26
369
23
336
 33
33
467
8
143
 53
53
746
113
366
 30
30
420
20
325
 21
21
299
68
152
 62
62
879
56
730
 27
27
386
22
295
 31
31
444
24
271
 40
40
563
22
398
 19
19
269
20
202
 26
26
376
22
462
 53
53
747
40
676
 25
25
357
27
308
 35
35
499
20
401
 100
100
1420
20
703
 16
16
222
14
143
 66
66
935
30
663
 43
43
617
21
733
 12
12
172
55
342
 74
74
1050
31
678
 46
46
648
22
1457
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 88.7
88.7
88.7
100 93.8
93.8
93.8
100 -
-
-
99 -
-
-
99 99.8
99.8
99.8
100 -
-
-
- -
-
-
100 99.6
99.6
99.8
100 -
-
-
- -
-
-
- 23.5
23.5
39.9
91 88.9
88.9
94.7
88.5 86.7
86.7
93.4
87 -
-
-
- 60.2
60.2
75.4
61 61.5
61.5
77.2
- 56.4
56.4
75.5
59 59.5
59.5
79.4
- -
-
-
- 24.5
24.5
44.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | LIMS1 - P48059 |
| 2 | AKT1 - P31749 |
| 3 | PARVA - Q9NVD7 |
| 4 | PXN - P49023 |
| 5 | ITGB1 - P05556 |
| 6 | PARVB - Q9HBI1 |
| 7 | LIMS2 - Q7Z4I7 |
| 8 | ITGB3 - P05106 |
| 9 | PARVG - Q9HBI0 |
| 10 | ITGB2 - P05107 |
| 11 | CASP9 - P55211 |
| 12 | SLC4A1AP - Q9BWU0 |
| 13 | CASP8 - Q14790 |
| 14 | ILKAP - Q9H0C8 |
| 15 | RICTOR - Q6R327 |
Regulation
Activation:
Stimulated rapidly but transiently by both cell fibronectin interactions, as well as by insulin, in a PI3-K-dependent manner, likely via the binding of PtdIns(3,4,5)P3 with a PH-like domain of ILK.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Akt1 (PKBa) | P31749 | S473 | RPHFPQFSYSASGTA | + |
| Akt2 (PKBb) | P31751 | S474 | RTHFPQFSYSASIRE | + |
| Akt3 (PKBg) | Q9Y243 | S472 | RPHFPQFSYSASGRE | + |
| Cofilin 1 | P23528 | S2 | ______ASGVAVSDG | |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| MRLC2 (MYL12B) | P19105 | S2 | _______SSKRTKTK | + |
| MRLC2 (MYL12B) | P19105 | S20 | KRPQRATSNVFAMFD | |
| MRLC2 (MYL12B) | P19105 | S3 | ______SSKRTKTKT | + |
| MRLC2 (MYL12B) | P19105 | T19 | KKRPQRATSNVFAMF | + |
| MYPT1 | O14974 | T500 | RLAYVAPTIPRRLAS | |
| MYPT1 | O14974 | T696 | ARQSRRSTQGVTLTD | |
| MYPT1 | O14974 | T710 | DLQEAEKTIGRSRST | |
| NACA | Q13765 | S43 | PELEEQDSTQATTQQ | |
| PPP1R14A (CPI 17) | Q96A00 | T38 | QKRHARVTVKYDRRE | - |
| PPP1R14B | Q96C90 | T57 | VRRQGKVTVKYDRKE | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
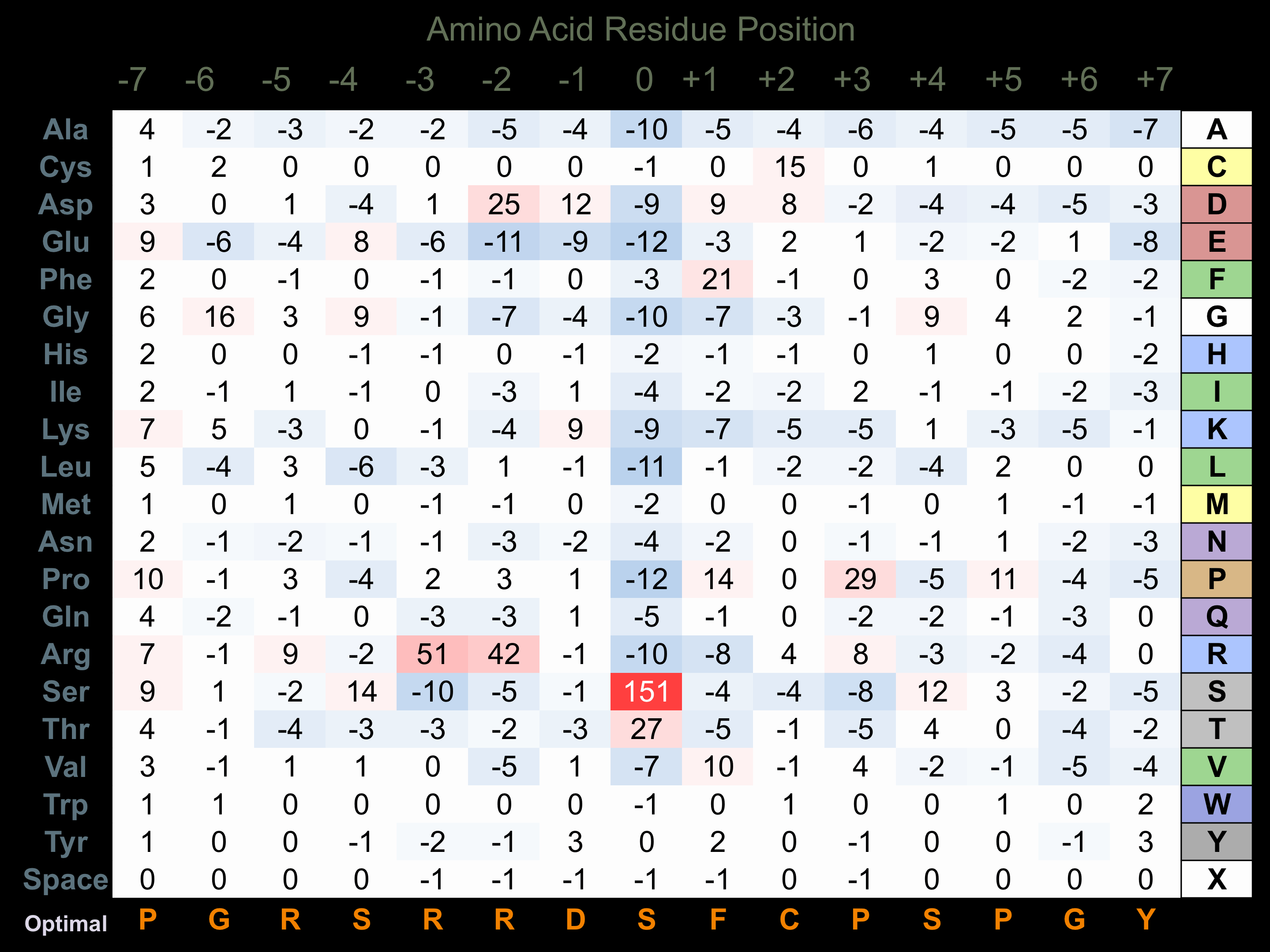
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer, skin disorders
Specific Diseases (Non-cancerous):
Scar contracture
Comments:
Scar contracture is a skin condition resulting from second or third degree burn, in which the skin around the burn pulls together and tightens, leading to scar formation and restriction in movement around the injured skin. Both ILK mRNA and protein expression has been observed in human dermal fibroblasts but absent in human epidermal cells in skin. In animal studies, mice with both V386G and T387G substitution mutations in the ILK gene displayed a reduction in the localization of ILK at focal adhesions resulting in defective migration of fibroblasts. Furthermore, human cells transfected with a kinase-deficient ILK protein (with an E359K substitution mutation) displayed a reduced capacity for the formation of the collagen lattice, an essential component of skin architecture. ILK expression has been observed in scar tissue during the first 6 months after formation, and it is hypothesized to have a role in the promotion of vascularization by regulating the expression of KDR and FLT-1 during the early stages of scar formation. A262V substitution in ILK is associated with dilated cardiomyopathy.
Specific Cancer Types:
Scar contracture
Comments:
ILK appears to be a oncoprotein (OP), but is relatively low rate of mutation in human cancers supports its assignment as a tumour requiring protein (TRP). It has been implicated as an oncoprotein due to its ability to activate Akt isoforms, although it has weak phosphotransferase activity that is stimulated in vitro with manganese.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Colorectal adenocarcinomas (early onset) (%CFC= +149, p<0.104); Large B-cell lymphomas (%CFC= +96, p<0.0008); Prostate cancer - metastatic (%CFC= -46, p<0.0001); Skin melanomas - malignant (%CFC= +199, p<0.0001); T-cell prolymphocytic leukemia (%CFC= +112, p<0.053); and Uterine leiomyomas (%CFC= -60, p<0.0008).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. The mutation H99D can modulate interactions with LIMS1. An E359K mutation can inactivate ILK.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24470 diverse cancer specimens. This rate is only -34 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.36 % in 1052 large intestine cancers tested.
Frequency of Mutated Sites:
None > 3 in 19,725 cancer specimens
Comments:
Only 6 deletions ( five L71fs*26), and no insertions or complex mutations are noted on the COSMIC website.

