Nomenclature
Short Name:
PAK1
Full Name:
Serine-threonine-protein kinase PAK 1
Alias:
- ADRB2
- Alpha-PAK
- MUK2
- P21 protein (Cdc42/Rac)-activated kinase 1
- P65-PAK
- STE20, yeast; P68-PAK; PAK 1; PAK-1; PAKA; Protein kinase MUK2;
- CDC42,RAC effector kinase PAK-A
- CDC42/RAC effector kinase PAK-A
- EC 2.7.11.1
- Kinase PAK1
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE20
SubFamily:
PAKA
Specific Links
Structure
Mol. Mass (Da):
60647
# Amino Acids:
545
# mRNA Isoforms:
2
mRNA Isoforms:
61,632 Da (553 AA; Q13153-2); 60,647 Da (545 AA; Q13153)
4D Structure:
Homodimer in its autoinhibited state. Active as monomer. Interacts tightly with GTP-bound but not GDP-bound CDC42/P21 and RAC1. Binds to the caspase-cleaved p110 isoform of CDC2L1 and CDC2L2, p110C, but not the full-length proteins. Component of cytoplasmic complexes, which also contain PXN, ARHGEF6 and GIT1. Interacts with ARHGEF7. Also interacts with CRIPAK. Interacts with NISCH By similarity. Probably found in a ternary complex composed of DSCAM, PAK1 and RAC1. Interacts with DSCAM (via cytoplasmic domain); the interaction is direct and enhanced in presence of RAC1. Interacts with SCRIB
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
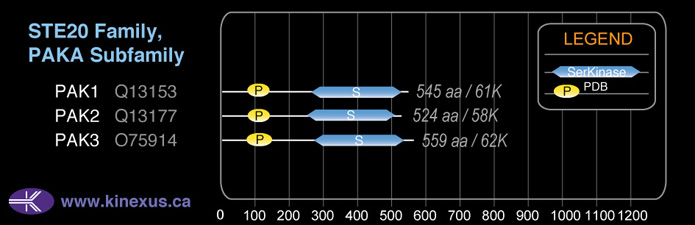
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K29, K256, K420, K513.
Serine phosphorylated:
S21-, S110, S115, S133, S137, S139, S144+, S149, S155, S156, S174, S199+, S204+, S220, S223, S262, S422+, S491, S528, S529.
Threonine phosphorylated:
T20, T84-, T109-, T136, T146, T185, T197, T212-, T214, T219, T225, T229, T230, T236, T415, T423+, T427, T531-.
Tyrosine phosphorylated:
Y131, Y142, Y153+, Y201+, Y285+, Y474.
Ubiquitinated:
K256, K420.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 45
45
1336
31
1205
 1.2
1.2
35
12
27
 6
6
168
14
141
 25
25
746
121
722
 27
27
809
34
520
 4
4
117
63
299
 13
13
389
43
530
 87
87
2597
48
5556
 18
18
526
10
397
 5
5
156
126
146
 3
3
80
37
95
 14
14
408
160
476
 3
3
76
25
106
 2
2
67
11
52
 4
4
134
21
165
 1
1
30
19
24
 7
7
206
470
1027
 3
3
92
22
82
 6
6
167
120
208
 21
21
622
112
531
 5
5
148
35
175
 5
5
162
35
204
 7
7
214
23
125
 5
5
156
23
120
 6
6
181
34
200
 24
24
703
79
673
 3
3
78
28
87
 5
5
147
23
124
 4
4
124
23
100
 6
6
182
42
148
 28
28
842
18
609
 100
100
2978
31
6391
 8
8
245
76
384
 27
27
809
83
742
 8
8
252
48
306
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 94.3
94.3
95.2
93.5 99.6
99.6
99.6
100 -
-
-
99 -
-
-
- 96.7
96.7
97.1
97 -
-
-
- 97.6
97.6
98.7
99 98.9
98.9
99.1
99 -
-
-
- 78.7
78.7
85.3
- 94.5
94.5
96.9
95 80.7
80.7
88.3
74 82
82
87.9
88 -
-
-
- 32.7
32.7
50.9
71 -
-
-
- 54.7
54.7
68.7
62 -
-
-
- -
-
-
- -
-
-
- -
-
-
49 -
-
-
- 32.6
32.6
43.7
55 32.7
32.7
43.8
40
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC42 - P60953 |
| 2 | RAC1 - P63000 |
| 3 | NCK1 - P16333 |
| 4 | ARHGEF7 - Q14155 |
| 5 | RAF1 - P04049 |
| 6 | RHOU - Q7L0Q8 |
| 7 | PAK3 - O75914 |
| 8 | CDK11B - P21127 |
| 9 | NF2 - P35240 |
| 10 | CDK11A - Q9UQ88 |
| 11 | AKT1 - P31749 |
| 12 | LIMK1 - P53667 |
| 13 | NCK2 - O43639 |
| 14 | CDK5R1 - Q15078 |
| 15 | SYN1 - P17600 |
Regulation
Activation:
Activated by binding small G proteins. Binding of GTP-bound Cdc42 or Rac1 to the autoregulatory region releases monomers from the autoinhibited dimer, enables phosphorylation at Thr-423 and allows the kinase domain to adopt an active structure. Also activated by binding to GTP-bound Cdc42, independent of the phosphorylation state of Thr-423. Phosphorylation of Ser-144, Ser-149, Thr-423 increases phosphotransferase activity.
Inhibition:
Phosphorylation of Tyr-212 inhibits phosphotransferase activity.Phosphorylation of Ser-21 inhibits binding of Nck and PIX. Phosphorylation of Thr-84 by OXSR1 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AKT3 | Q9Y243 | S21 | APPMRNTSTMIGAGS | - |
| Akt1 | P31749 | S21 | APPMRNTSTMIGAGS | - |
| PAK1 | Q13153 | S21 | APPMRNTSTMIGAGS | - |
| PKG1 | Q13976 | S21 | APPMRNTSTMIGAGS | - |
| OSR1 | O95747 | T84 | LPSDFEHTIHVGFDA | - |
| PAK1 | Q13153 | S144 | SNSQKYMSFTDKSAE | + |
| Akt1 | P31749 | S199 | PRPEHTKSVYTRSVI | + |
| PAK1 | Q13153 | S199 | PRPEHTKSVYTRSVI | + |
| JAK2 | O60674 | Y201 | PEHTKSVYTRSVIEP | + |
| Akt1 | P31749 | S204 | TKSVYTRSVIEPLPV | + |
| PAK1 | Q13153 | S204 | TKSVYTRSVIEPLPV | + |
| CDK1 | P06493 | T212 | VIEPLPVTPTRDVAT | - |
| ERK2 | P28482 | T212 | VIEPLPVTPTRDVAT | - |
| CDK5 | Q00535 | T212 | VIEPLPVTPTRDVAT | - |
| PKACa | P17612 | T423 | PEQSKRSTMVGTPYW | + |
| PAK1 | Q13153 | T423 | PEQSKRSTMVGTPYW | + |
| PDK1 | O15530 | T423 | PEQSKRSTMVGTPYW | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ARHGDIA | P52565 | S101 | LESFKKQSFVLKEGV | |
| ARHGDIA | P52565 | S174 | KGMLARGSYSIKSRF | |
| ARHGEF2 | Q8TDA3 | S885 | PVDPRRRSLPAGDAL | |
| ARHGEF2 | Q92974 | S885 | PVDPRRRSLPAGDAL | |
| ARHGEF7 iso2 | Q14155-2 | T654 | EFASRKSTAALEEDA | |
| ARPC1B (p41-Arc) | O15143 | T21 | HAWNKDRTQIAICPN | ? |
| AurA (STK15) | O14965 | S342 | QETYKRISRVEFTFP | + |
| AurA (STK15) | O14965 | T288 | APSSRRTTLCGTLDY | + |
| B-Raf | P15056 | S446 | KTLGRRDSSDDWEIP | + |
| Bad | Q92934 | S134 | KGLPRPKSAGTATQM | - |
| Bad | Q92934 | S75 | EIRSRHSSYPAGTED | - |
| Bad | Q92934 | S99 | PFRGRSRSAPPNLWA | - |
| CtBP1 | Q13363 | S158 | REGTRVQSVEQIREV | |
| CTTN (Cortactin) | Q14247 | S405 | KTQTPPVSPAPQPTE | - |
| ELF3 | P78545 | S207 | GTGASRSSHSSDSGG | - |
| ERa (ESR1) | P03372 | S305 | IKRSKKNSLALSLTA | + |
| ERK3 (MAPK6) | Q16659 | S189 | YSHKGHLSEGLVTKW | + |
| ERK4 (MAPK4) | P31152 | S186 | YSHKGYLSEGLVTKW | + |
| FLNA | P21333 | S2152 | TRRRRAPSVANVGSH | |
| FLNA | P21333 | S2292 | FEDRKDGSCGVAYVV | |
| FLNA | P21333 | S2370 | AIDAKVHSPSGALEE | |
| Ga(z) | P19086 | S16 | EKEAARRSRRIDRHL | |
| Ga(z) | P19086 | Y154 | HLEDNAAYYLNDLER | |
| GIT1 | Q9Y2X7 | S508 | RRDRQAFSMYEPGSA | |
| GIT1 | Q9Y2X7 | S700 | PALEPVRSSLRLLNA | |
| H3.1 | P68431 | S11 | TKQTARKSTGGKAPR | + |
| hnRNP E1 | Q15365 | T127 | IKEIRESTGAQVQVA | |
| hnRNP E1 | Q15365 | T60 | NCPERIITLTGPTNA | |
| ILK | Q13418 | S246 | CPRLRIFSHPNVLPV | ? |
| ILK | Q13418 | T173 | DTFWKGTTRTRPRNG | ? |
| LC8 | P63167 | S88 | VAILLFKSG______ | |
| LIMK1 | P53667 | T508 | PDRKKRYTVVGNPYW | + |
| MEK1 (MAP2K1) | Q02750 | S298 | RTPGRPLSSYGMDSR | + |
| MEKK1 (MAP3K1) | Q13233 | S67 | RQLRKVRSVELDQLP | - |
| Merlin | P35240 | S518 | DTDMKRLSMEIEKEK | |
| MRLC1 (MYL9) | P24844 | S20 | KRPQSATSNVFAMFD | |
| MRLC1 (MYL9) | P24844 | T19 | KKRPQRATSNVFAMF | |
| MYLK1 (smMLCK) | Q15746 | S1772 | SGLSGRKSSTGSPTS | |
| MYPT1 | O14974 | T696 | ARQSRRSTQGVTLTD | |
| Net1 | Q7Z628 | S152 | PTPAKRRSSALWSEM | |
| Net1 | Q7Z628 | S153 | TPAKRRSSALWSEML | |
| Net1 | Q7Z628 | S539 | LTAQRRASTVSSVTQ | |
| NRIF3 | Q13352 | S28 | SKITRKKSVITYSPT | + |
| PA2G4 | Q9UQ80 | T261 | QYGLKMKTSRAFFSE | - |
| PAK1 | Q13153 | S144 | SNSQKYMSFTDKSAE | + |
| PAK1 | Q13153 | S199 | PRPEHTKSVYTRSVI | + |
| PAK1 | Q13153 | S204 | TKSVYTRSVIEPLPV | + |
| PAK1 | Q13153 | S21 | APPMRNTSTMIGAGS | - |
| PAK1 | Q13153 | T423 | PEQSKRSTMVGTPYW | + |
| PGAM1 | P18669 | S118 | QVKIWRRSYDVPPPP | |
| PGAM1 | P18669 | S23 | WNLENRFSGWYDADL | |
| PGM1 | P36871 | T467 | SANDKVYTVEKADNF | |
| Plk1 (PLK) | P53350 | S49 | EVLVDPRSRRRYVRG | + |
| PPP1R14A (CPI 17) | Q96A00 | T38 | QKRHARVTVKYDRRE | - |
| PXN | P49023 | S272 | ELDELMASLSDFKFM | ? |
| Raf1 | P04049 | S338 | RPRGQRDSSYYWEIE | + |
| Raf1 | P04049 | S339 | PRGQRDSSYYWEIEA | + |
| SHARP | Q96T58 | S3486 | QPAPKQDSSPHLTSQ | |
| SHARP | Q96T58 | T3568 | EGVARRMTVETDYCL | |
| SNAI1 | O95863 | S246 | QACARTFSRMSLLHK | |
| STAT5A | P42229 | S780 | DSLDSRLSPPAGLFT | - |
| STMN1 | P16949 | S16 | KELEKRASGQAFELI | |
| STMN1 | P16949 | S38 | SVPEFPLSPPKKKDL | |
| SYN1 | P17600 | S551 | PAARPPASPSPQRQA | |
| SYN1 | P17600 | S605 | AGPTRQASQAGPVPR | |
| SYN1 | P17600 | S9 | NYLRRRLSDSNFMAN | |
| TBCB | Q99426 | S128 | VRSFLKRSKLGRYNE | |
| TBCB | Q99426 | S65 | GVDDKFYSKLDQEDA | |
| Vimentin | P08670 | S26 | GTASRPSSSRSYVTT | |
| Vimentin | P08670 | S39 | TTSTRTYSLGSALRP | - |
| Vimentin | P08670 | S51 | LRPSTSRSLYASSPG | |
| Vimentin | P08670 | S56 | SRSLYASSPGGVYAT | |
| Vimentin | P08670 | S66 | GVYATRSSAVRLRSS | - |
| Vimentin | P08670 | S73 | SAVRLRSSVPGVRLL |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
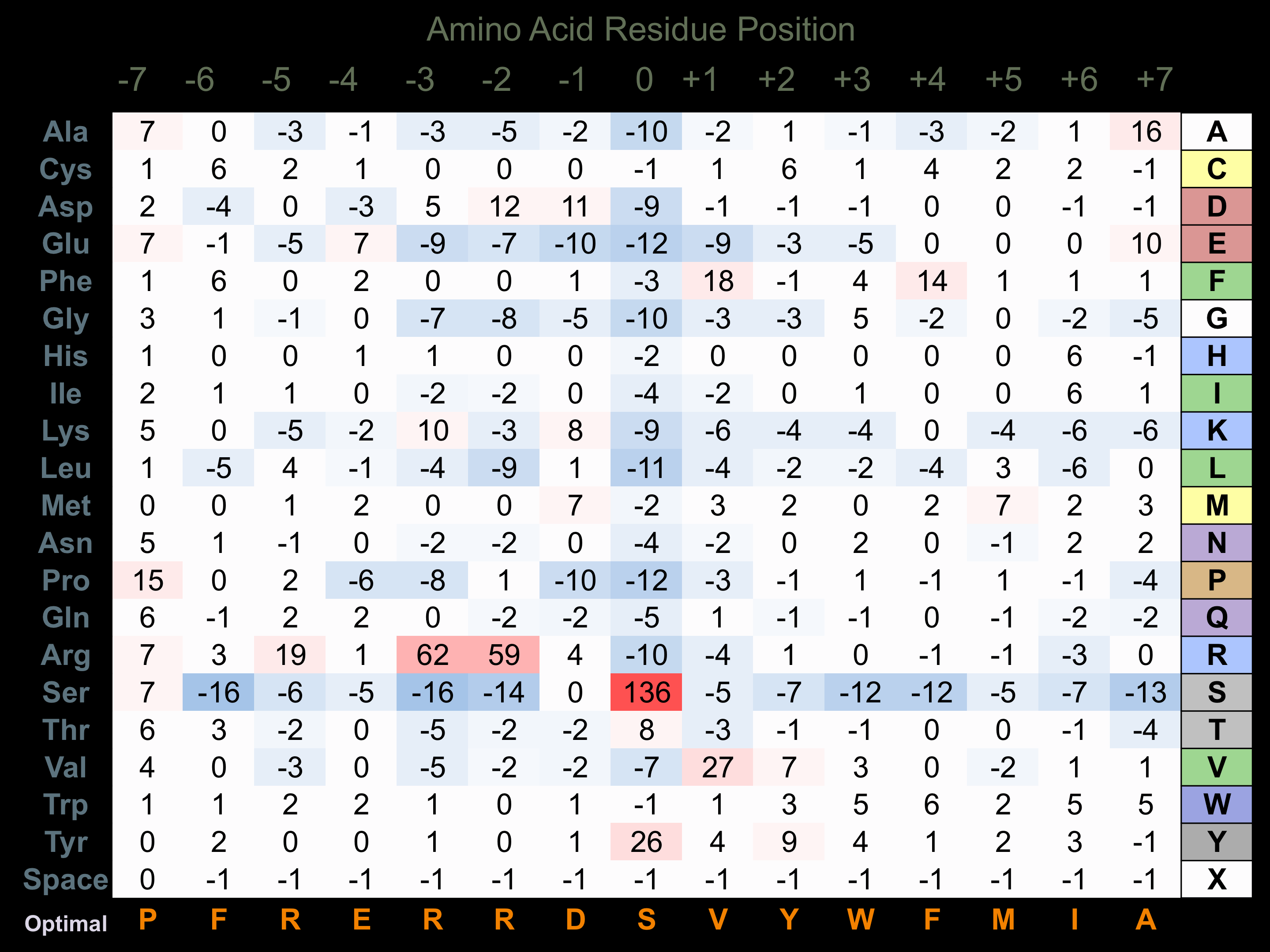
Matrix Type:
Experimentally derived from alignment of 97 known protein substrate phosphosites and 95 peptides phosphorylated by recombinant PAK1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Melanomas; Colon cancers
Comments:
PAK1 may be a therapeutic target in B-Raf wild-type melanoma. Knockdown of PAK1 has been shown to inhibit the proliferation of mutant KRAS colon cancer cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +45, p<0.006); Classical Hodgkin lymphomas (%CFC= +105, p<0.009); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +864, p<0.0004); Large B-cell lymphomas (%CFC= +99, p<0.013); Lung adenocarcinomas (%CFC= +126, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= +123, p<0.094); and Prostate cancer - metastatic (%CFC= -65, p<0.0001). The COSMIC website notes an up-regulated expression score for PAK1 in diverse human cancers of 496, which is 1.1-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 92 for this protein kinase in human cancers was 1.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25366 diverse cancer specimens. This rate is only -4 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.4 % in 1093 large intestine cancers tested; 0.27 % in 805 skin cancers tested; 0.12 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,632 cancer specimens
Comments:
Only 1 insertion, and no deletions or complex mutations are noted on the COSMIC website.

