Nomenclature
Short Name:
JAK2
Full Name:
Tyrosine-protein kinase JAK2
Alias:
- EC 2.7.10.2
- JAK-2
- Janus kinase 2
- JTK10
- Kinase Jak2
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
JakA
SubFamily:
NA
Specific Links
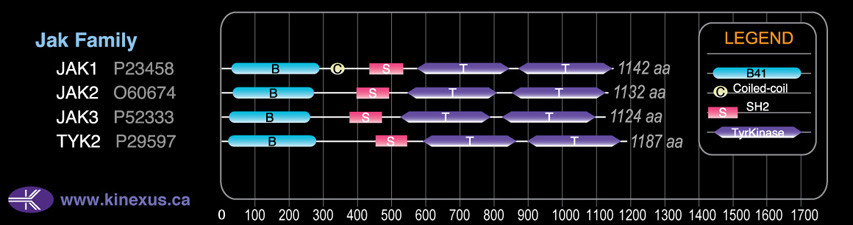
Structure
Mol. Mass (Da):
130,674
# Amino Acids:
1132
# mRNA Isoforms:
1
mRNA Isoforms:
130,674 Da (1132 AA; O60674)
4D Structure:
Interacts with EPOR, SIRPA and SH2B1 By similarity. Interacts with IL23R, SKB1 and STAM2.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K1069.
Serine phosphorylated:
S523-, S904, S936.
Sumoylated:
K167, K630, K912, K914, K991, K1011.
Threonine phosphorylated:
T522.
Tyrosine phosphorylated:
Y119-, Y201, Y206, Y221, Y317, Y382, Y423, Y435, Y570+, Y613+, Y637, Y790, Y813-, Y868, Y913-, Y918, Y931, Y934, Y940, Y956, Y966+, Y972+, Y1007+, Y1008+.
Ubiquitinated:
K207, K762.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1120
35
1056
 2
2
20
16
12
 9
9
104
26
86
 16
16
177
130
310
 70
70
786
30
538
 4
4
42
86
77
 12
12
139
41
345
 29
29
326
56
441
 39
39
440
17
368
 5
5
53
99
54
 4
4
45
47
51
 53
53
595
174
550
 13
13
144
48
147
 4
4
41
12
27
 4
4
44
38
59
 2
2
18
20
13
 3
3
31
214
45
 19
19
216
34
794
 12
12
133
101
136
 60
60
672
132
631
 9
9
100
39
97
 15
15
168
42
173
 15
15
163
28
140
 10
10
110
35
102
 8
8
95
39
101
 35
35
389
87
478
 9
9
106
51
107
 7
7
78
35
78
 11
11
124
35
116
 3
3
31
28
27
 58
58
655
24
599
 34
34
377
41
426
 4
4
44
85
60
 72
72
801
78
622
 5
5
52
48
53
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
99.7
100 99.3
99.3
99.7
99 -
-
-
95 -
-
-
95 94.5
94.5
97.5
94.5 -
-
-
- 93.5
93.5
97.4
94 94.6
94.6
97.3
95 -
-
-
- 78.1
78.1
81.4
- 87
87
93.4
87 77.7
77.7
89.1
78 42.1
42.1
60.7
70 -
-
-
- -
-
-
24 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CSF2RB - P32927 |
| 2 | EPOR - P19235 |
| 3 | GHR - P10912 |
| 4 | IRS1 - P35568 |
| 5 | STAT5B - P51692 |
| 6 | EGFR - P00533 |
| 7 | PTPN6 - P29350 |
| 8 | STAT3 - P40763 |
| 9 | INSR - P06213 |
| 10 | RAF1 - P04049 |
| 11 | CCR5 - P51681 |
| 12 | IL12RB2 - Q99665 |
| 13 | VAV1 - P15498 |
| 14 | PIK3R1 - P27986 |
| 15 | CTLA4 - P16410 |
Regulation
Activation:
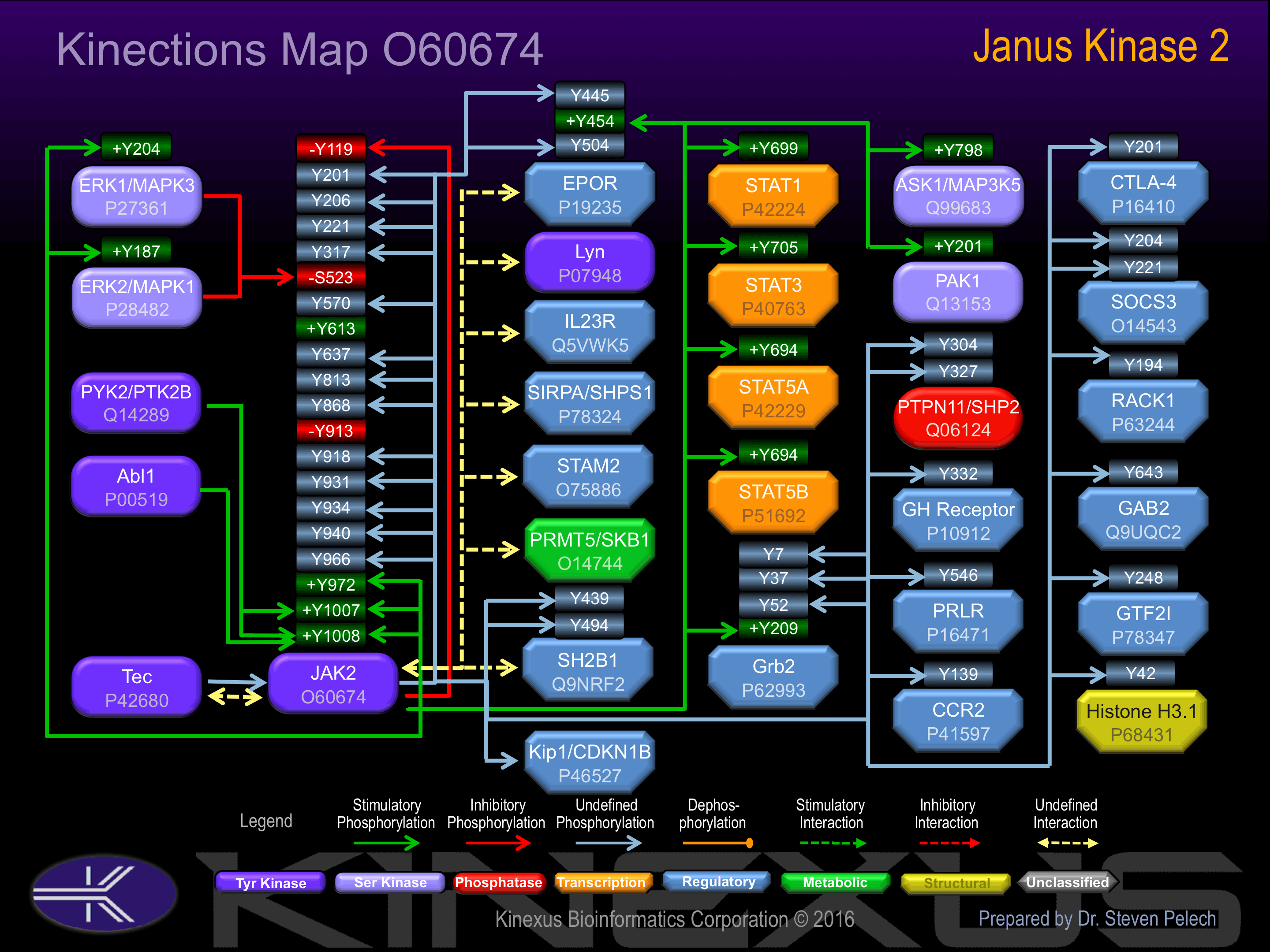
Activated by phosphorylation at Tyr-613, and Tyr-972, and Tyr-1008.
Inhibition:
Inhibited by phosphorylation at Tyr-119, Ser-523 and Tyr-913.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| JAK2 | O60674 | Y119 | VLYRIRFYFPRWYCS | - |
| JAK2 | O60674 | Y201 | DQTPLAIYNSISYKT | |
| JAK2 | O60674 | Y206 | AIYNSISYKTFLPKC | |
| JAK2 | O60674 | Y221 | IRAKIQDYHILTRKR | |
| JAK2 | O60674 | Y317 | TEQDLQLYCDFPNII | |
| ERK2 | P28482 | S523 | GVSDVPTSPTLQRPT | - |
| ERK1 | P27361 | S523 | GVSDVPTSPTLQRPT | - |
| JAK2 | O60674 | Y570 | VRREVGDYGQLHETE | |
| JAK2 | O60674 | Y637 | KFGSLDTYLKKNKNC | |
| JAK2 | O60674 | Y813 | NSLFTPDYELLTEND | ? |
| JAK2 | O60674 | Y868 | GSVEMCRYDPLQDNT | |
| JAK2 | O60674 | Y913 | QHDNIVKYKGVCYSA | - |
| JAK2 | O60674 | Y918 | VKYKGVCYSAGRRNL | |
| JAK2 | O60674 | Y931 | NLKLIMEYLPYGSLR | |
| JAK2 | O60674 | Y934 | LIMEYLPYGSLRDYL | |
| JAK2 | O60674 | Y940 | PYGSLRDYLQKHKER | |
| JAK2 | O60674 | Y966 | QICKGMEYLGTKRYI | |
| JAK2 | O60674 | Y972 | EYLGTKRYIHRDLAT | + |
| PYK2 | Q14289 | Y1007 | VLPQDKEYYKVKEPG | + |
| JAK2 | O60674 | Y1007 | VLPQDKEYYKVKEPG | + |
| PYK2 | Q14289 | Y1008 | LPQDKEYYKVKEPGE | + |
| JAK2 | O60674 | Y1008 | LPQDKEYYKVKEPGE | + |
| ABL | P00519 | Y1008 | LPQDKEYYKVKEPGE | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ASK1 (MAP3K5) | Q99683 | Y798 | IPERDSRYSQPLHEE | |
| CCR2 | P41597 | Y139 | ILLTIDRYLAIVHAV | |
| CTLA-4 | P16410 | Y201 | SPLTTGVYVKMPPTE | |
| EpoR | P19235 | Y445 | PPHLKYLYLVVSDSG | |
| EpoR | P19235 | Y454 | PTPPHLKYLYLVVSD | + |
| EpoR | P19235 | Y504 | AEPLPPSYVACS___ | ? |
| ERK1 (MAPK3) | P27361 | Y204 | HTGFLTEYVATRWYR | + |
| ERK2 (MAPK1) | P28482 | Y187 | HTGFLTEYVATRWYR | + |
| GAB2 | Q9UQC2 | Y643 | TSDEKVDYVQVDKEK | |
| GH receptor | P10912 | Y332 | ILAIHDSYKPEFHSD | |
| Grb2 | P62993 | Y209 | TGMFPRNYVTPVNRN | + |
| Grb2 | P62993 | Y37 | EECDQNWYKAELNGK | ? |
| Grb2 | P62993 | Y52 | DGFIPKNYIEMKPHP | ? |
| Grb2 | P62993 | Y7 | _MEAIAKYDFKATAD | ? |
| GTF2I | P78347 | Y248 | EESEDPDYYQYNIQG | |
| Histone H3 | P68431 | Y42 | GVkkPHrYRPGTVAL | ? |
| JAK2 | O60674 | Y1007 | VLPQDKEYYKVKEPG | + |
| JAK2 | O60674 | Y1008 | LPQDKEYYKVKEPGE | + |
| JAK2 | O60674 | Y119 | VLYRIRFYFPRWYCS | - |
| JAK2 | O60674 | Y201 | DQTPLAIYNSISYKT | |
| JAK2 | O60674 | Y206 | AIYNSISYKTFLPKC | |
| JAK2 | O60674 | Y221 | IRAKIQDYHILTRKR | |
| JAK2 | O60674 | Y317 | TEQDLQLYCDFPNII | |
| JAK2 | O60674 | Y570 | VRREVGDYGQLHETE | |
| JAK2 | O60674 | Y637 | KFGSLDTYLKKNKNC | |
| JAK2 | O60674 | Y813 | NSLFTPDYELLTEND | ? |
| JAK2 | O60674 | Y868 | GSVEMCRYDPLQDNT | |
| JAK2 | O60674 | Y913 | QHDNIVKYKGVCYSA | - |
| JAK2 | O60674 | Y918 | VKYKGVCYSAGRRNL | |
| JAK2 | O60674 | Y931 | NLKLIMEYLPYGSLR | |
| JAK2 | O60674 | Y934 | LIMEYLPYGSLRDYL | |
| JAK2 | O60674 | Y940 | PYGSLRDYLQKHKER | |
| JAK2 | O60674 | Y966 | QICKGMEYLGTKRYI | |
| JAK2 | O60674 | Y972 | EYLGTKRYIHRDLAT | + |
| PAK1 | Q13153 | Y201 | PEHTKSVYTRSVIEP | + |
| PRLR | P16471 | Y546 | TPENNKEYAKVSGVM | |
| PTPN11 (SHP2) | Q06124 | Y304 | PNEPVSDYINANIIM | |
| PTPN11 (SHP2) | Q06124 | Y327 | NSKPKKSYIATQGCL | |
| RACK1 | P63244 | Y194 | NHIGHTGYLNTVTVS | |
| SH2-Bb | Q9NRF2 | Y439 | DRLSQGAYGGLSDRP | |
| SH2-Bb | Q9NRF2 | Y494 | VHPLSAPYPPLDTPE | |
| SOCS3 | O14543 | Y204 | VNGHLDSYEKVTQLP | |
| SOCS3 | O14543 | Y221 | IREFLDQYDAPL___ | |
| STAT1 | P42224 | Y701 | DGPKGTGYIKTELIS | + |
| STAT3 | P40763 | Y705 | DPGSAAPYLKTKFIC | + |
| STAT5A | P42229 | Y694 | LAKAVDGYVKPQIKQ | + |
| STAT5B | P51692 | Y699 | TAKAVDGYVKPQIKQ | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
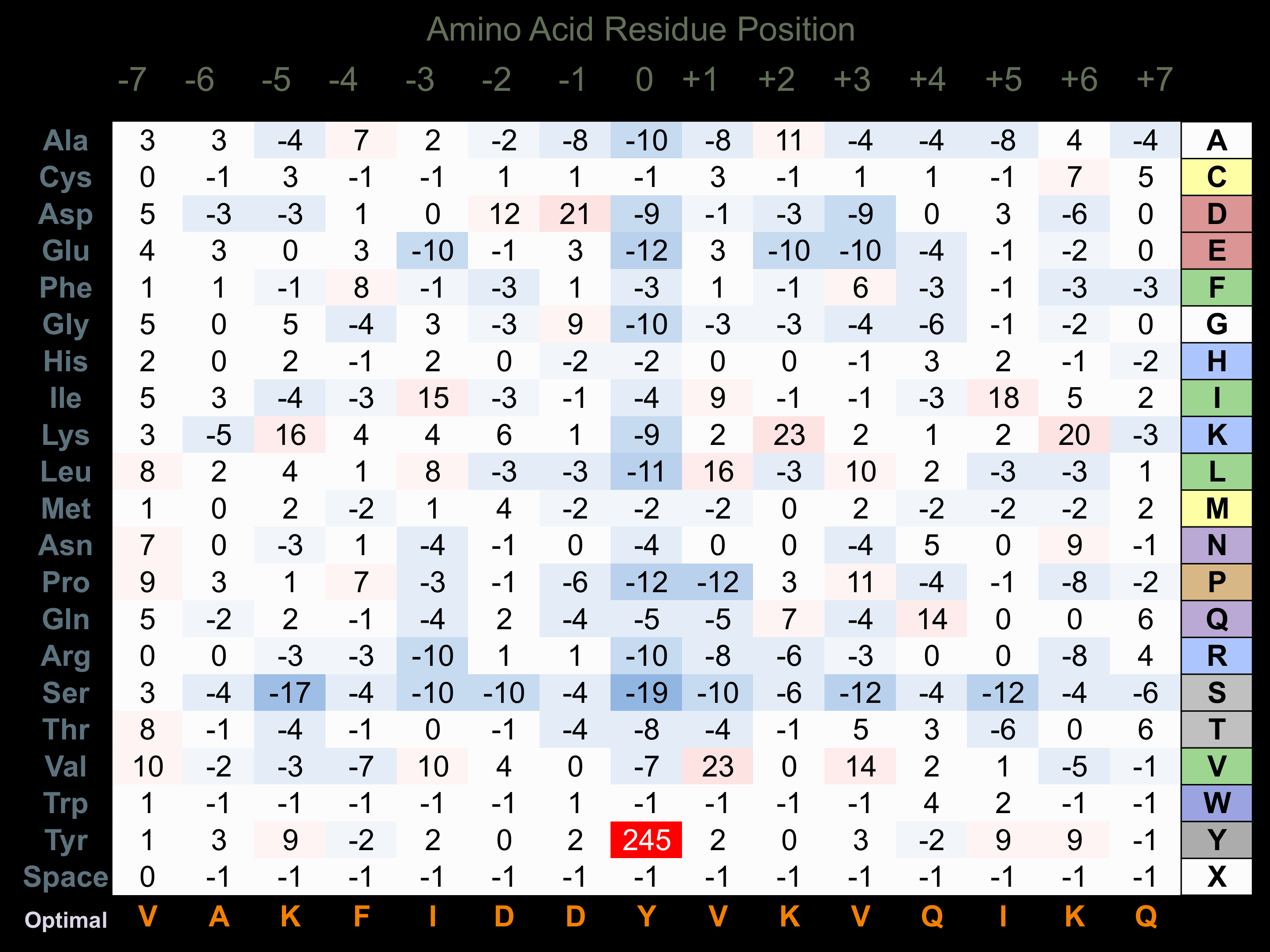
Matrix Type:
Experimentally derived from alignment of 85 known protein substrate phosphosites.
Domain #:
2
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, blood disorders
Specific Diseases (Non-cancerous):
Polycythemia vera; Polycythemia; Myelofibrosis; Thrombocytosis; Erythrocytosis; Thrombocythemia 3; Essential thrombocythemia; Hepatic vein thrombosis; Central retinal vein occlusion; Acquired polycythemia; Thrombocythemia 1; Erythrocytosis, somatic; Myelofibrosis with myeloid metaplasia, somatic; Myelofibrosis, somatic; JAK2-related Budd-Chiari syndrome
Comments:
Polycythemia vera, thrombocythemia, and myelofibrosis are classified as clonal myeloproliferative diseases that arise from a multipotent progenitor cell type. These often rare diseases are characterized by excess production of abnormal cells, displacing healthy cells. Polycythemia vera is a disease characterized by increased numbers of circulating red blood cells, often combined with increased white blood cells and platelets. The increased cell number results in thickening of the blood leading to a higher risk of blood clots. Affected individuals also have a predisposition for deep vein thrombosis (DVT), a blood clot that occurs in the deep veins of the arms and legs. Thrombocythemia 3 (THCYT3) is characterized by excessive platelet production and increased circulating platelets, and while it may be symptom-less, it can increase the risk of thrombosis. Essential Thrombocythemia is another myeloproliferative causing increased production of white blood cells, red blood cells, and platelets. Thrombocythemia 1 (ET) is a rare disorder related to polycythemia and polycythemia vera. ET can affect the bone and bone marrow. It can manifest with symptoms including pulmonary hypertension (increased blood pressure in the right side of the heart and in the lungs), numbness, and stroke. >The V617F mutation in JAK2 is associated thrombocythemia 3, myelofibrosis. and susceptibility to Budd-Chiari syndrome. JAK2-Related Budd-Chiari Syndrome (Jak2-Related BDCHS) is characterized by JAK2 caused occlusion of hepatic veins that drain the liver, resulting in liver damage. Clinical observations for JAK2-Related Budd-Chiari Syndrome involve hepatomegaly, right upper quadrant pain and abdominal ascites. The V617F mutation can also result in hypersensitivity to cytokines. Mutations in Y931 or V617F can make JAK2 immune to JAK inhibitors like INCB018424. Additionally, some patients with thrombocythemia displayed a V617I substitution mutation in the JAK2 gene correlated with megakaryocyte hyperplasia in the bone marrow. >Other mutations in the JAK2 gene have also been associated with the above diseases. For example, a K539L substitution mutation was observed in patients with polycythemia vera or idiopathic erythrocytosis, resulting in a distinctive bone marrow morphology and reduced serum EPO levels. Over-activation of STAT5 has been linked to the TEL/JAK2 fusion gene in myelo- and lymphoproliferative diseases. In animal studies, mice transplanted with bone marrow transduced with the TEL/JAK2 fusion gene rapidly develop myelo- and lymphoproliferative syndrome resulting in death shortly afterwards. Somatic mutations in the JAK2 gene were reported in 18% (16 of 88) of patients with Down-syndrome-associated acute lymphoblastic leukemia (ALL), whereas only 1 of 109 patients without the disease had a mutation in the gene. >Myelofibrosis (MYELOF) is a rare disease characterized by the replacement of the bone marrow with fibrous scar tissue, leading to anemia and the accumulation of blood in abnormal locations, such as the liver and spleen, which causes these organs to swell. Polycythemia vera is a disease characterized by increased numbers of circulating red blood cells, often combined with increased white blood cells and platelets. The increased cell number results in thickening of the blood leading to a higher risk of blood clots. Affected individuals also have a predisposition for deep vein thrombosis (DVT), a blood clot that occurs in the deep veins of the arms and legs. Thrombocythemia 3 (THCYT3) is characterized by excessive platelet production and increased circulating platelets, and while it may be symptom-less, it can increase the risk of thrombosis. Essential Thrombocythemia is another myeloproliferative causing increased production of white blood cells, red blood cells, and platelets. Thrombocythemia 1 (ET) is a rare disorder related to polycythemia and polycythemia vera. ET can affect the bone and bone marrow. It can manifest with symptoms including pulmonary hypertension (increased blood pressure in the right side of the heart and in the lungs), numbness, and stroke. Myelofibrosis (MYELOF) is a rare disease characterized by the replacement of the bone marrow with fibrous scar tissue, leading to anemia and the accumulation of blood in abnormal locations, such as the liver and spleen, which causes these organs to swell.
Specific Cancer Types:
Chronic myeloproliferative disease; Acute myeloid leukemias (AML), adult; Acute myeloid leukemias (AML); Primary mediastinal large B-cell lymphomas; Core binding factor Acute myeloid leukemias (AML); 3q21q26 syndrome; Acute erythroid leukemias; leukemias, acute myeloid, reduced survival in; leukemias, acute myeloid, somatic; Myeloid leukemias, acute, M4/m4eo subtype, somatic; Myeloproliferative disorder
Comments:
JAK2 is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. JAK2 signalling has been implicated in in a wide range of human hematologic malignancies, and this is typically accompanied by the over-activation of the STAT5 transcription factor, which is a substrate. Over-activation of STAT5 has been linked to the TEL/JAK2 fusion gene in myelo- and lymphoproliferative diseases. In animal studies, mice transplanted with bone marrow transduced with the TEL/JAK2 fusion gene rapidly develop myelo- and lymphoproliferative syndrome resulting in death shortly afterwards. Somatic mutations in the JAK2 gene were reported in 18% (16 of 88) of patients with Down-syndrome-associated acute lymphoblastic leukemia (ALL), whereas only 1 of 109 patients without the disease had a mutation in the gene. Acute myeloid leukemia (AML) is a white blood cell cancer (leukemia) where abnormal white blood cells are overproduced in the bone marrow leading to repressed production of healthy blood cells. The rare disorder Acute Myeloid Leukemia, Adult has a relation to acute leukemia and myeldysplastic syndromes. >These myeloproliferative diseases are usually associated with the loss of heterozygosity (LOH) on chromosome 9p, indicating that this region contains a mutation that contributes to the pathologies of these diseases. A high percentage of patients with these myeloproliferative diseases have a dominant gain-of-function substitution mutation, V617F, in the JAK2 gene sequence. For example, the V617F mutation was observed in 40 of 45 polycythemia vera patients. The mutation was observed to cause constitutive tyrosine phosphotransferase activity that promoted erythrocytosis in a mouse model. K607N and V617F mutations are linked to acute myelogenous leukemia (AML). Three distinct mutations were observed in patients with acute lymphoblastic leukemia (ALL), each occurring at the highly conserved R683 residues of the protein; R683G, R683S, R683K. Analysis of the mutant proteins revealed constitutive JAK2 activity and cytokine-independent cell growth, indicating gain-of-function mutations. In a patient with early pre-B acute lymphoid leukemia a t(9;12)(p24;p13) translocation was identified, and in a patient with atypical chronic myelogenous leukemia (CML) a t(9;15)(p24;q15;p13) translocation was observed. Both of these translocations involved the ETV6 gene (located at 12p13) and the JAK2 gene (located at 9p24) and produced a chimeric protein with the oligomerization domain of the ETV6 protein and the tyrosine kinase domain of JAK2 with constitutive phosphotransferase activity that produced cytokine independent proliferative ability onto the affected cells. In 2 out of 113 patients with acute myelogenous leukemia (AML) the substitution mutation V617F was found in the JAK2 gene. In contrast, JAK2 mutations were not observed in 94 ductal breat carcinomas, 104 colorectal carcinomas, or 217 small cell lung cancer samples. In 1 of 113 patients with AML a K607N substitution was observed in the JAK2 gene, which is located in a conserved residue in the pseudokinase domain of the protein. Myelofibrosis with Myeloid Metaplasia, Somatic is another rare cancer disease related to the myelofibrosis and polycythemia disorders.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer (%CFC= +55, p<0.072); and Oral squamous cell carcinomas (OSCC) (%CFC= +63, p<0.034). The COSMIC website notes an up-regulated expression score for JAK2 in diverse human cancers of 342, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 11 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 2.8 % in 135427 diverse cancer specimens. This rate is 37-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 3.63 % in 103660 haematopoietic and lymphoid cancers tested; 0.38 % in 1992 large intestine cancers tested; 0.17 % in 792 endometrium cancers tested; 0.13 % in 1115 stomach cancers tested; 0.11 % in 3383 lung cancers tested; 0.11 % in 1306 skin cancers tested; 0.08 % in 1361 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: V617F (39,717); R683G (45).
Comments:
In human tumours, the point mutations are mostly at amino acid residue 617, and deletions, insertions and complex mutations primarily around amino acid residues 536-556.