Nomenclature
Short Name:
LATS1
Full Name:
Serine-threonine protein kinase LATS1
Alias:
- EC 2.7.11.1
- Large tumor suppressor 1
- LATS, large tumor suppressor, 1
- WARTS
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
NDR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
126,870
# Amino Acids:
1130
# mRNA Isoforms:
2
mRNA Isoforms:
126,870 Da (1130 AA; O95835); 76,193 Da (690 AA; O95835-2)
4D Structure:
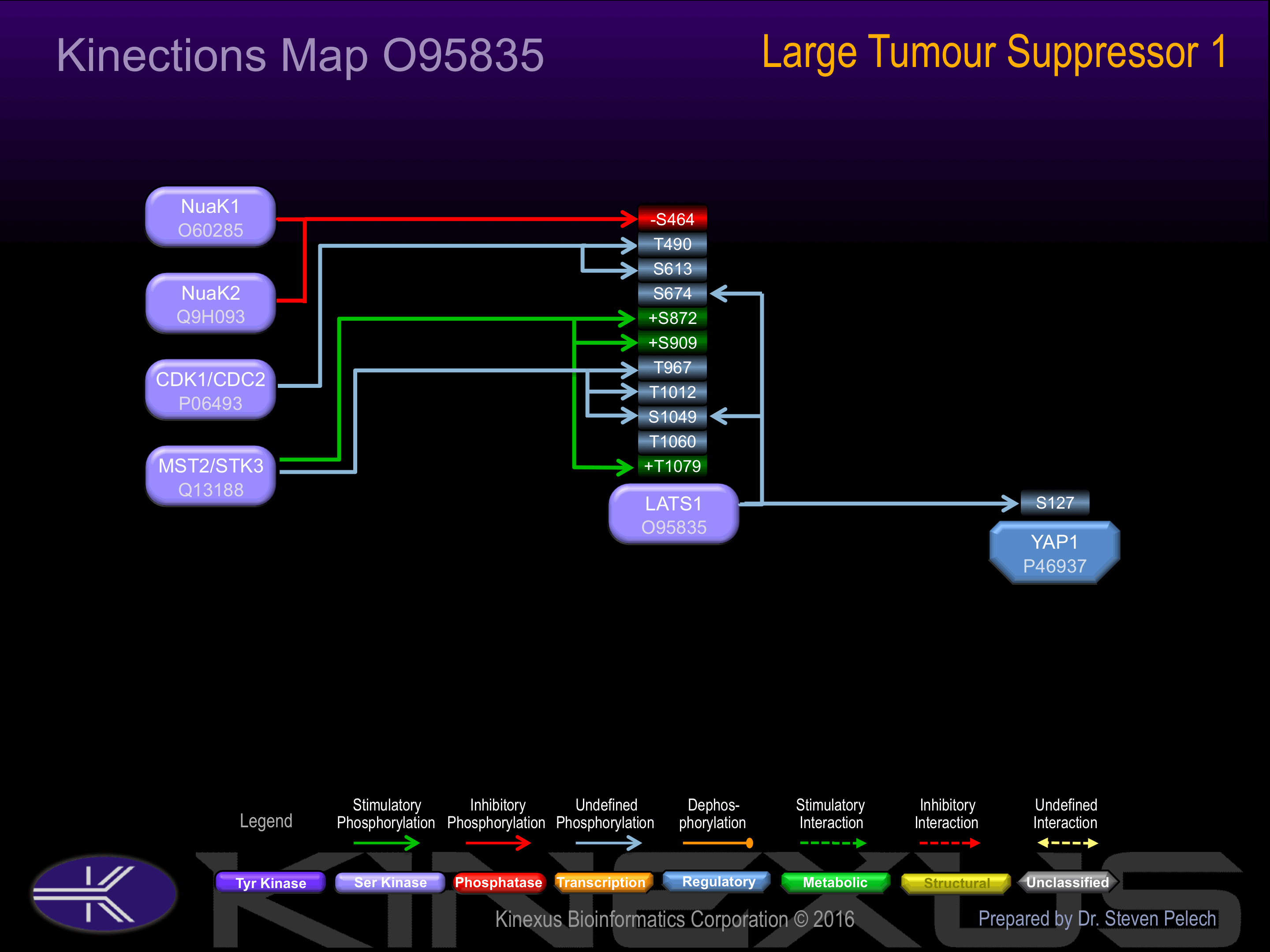
Complexes with CDK1 in early mitosis. LATS1-associated CDK1 has no mitotic cyclin partner and no apparent kinase activity. Binds phosphorylated ZYX, locating this protein to the mitotic spindle and suggesting a role for actin regulatory proteins during mitosis. Binds to and colocalizes with LIMK1 at the actomyosin contractile ring during cytokinesis. Interacts (via PPxY motif 2) with YAP1 (via WW domains). Interacts with MOBKL1A and MOBKL1B.
1D Structure:
Subfamily Alignment
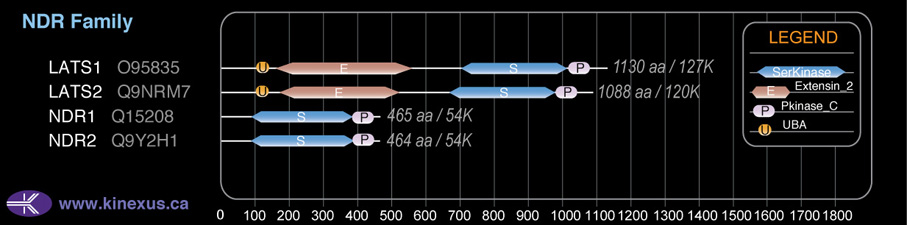
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 100 | 141 | UBA |
| 160 | 560 | Extensin_2 |
| 705 | 1010 | Pkinase |
| 1011 | 1090 | Pkinase_C |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S26, S27, S42, S171, S177, S181, S244, S278, S336, S462, S464-, S580, S613, S633, S635, S674, S792, S864, S872+, S909+, S1049, S1111.
Threonine phosphorylated:
T17, T246, T255, T261, T262, T490, T612, T616, T967, T1012, T1060, T1079+.
Tyrosine phosphorylated:
Y23, Y143, Y277, Y283, Y1076.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 94
94
1564
28
1499
 9
9
150
10
199
 10
10
158
7
90
 22
22
373
90
1527
 29
29
476
28
244
 0.5
0.5
8
55
6
 3
3
42
35
102
 52
52
861
19
1179
 25
25
423
10
280
 4
4
66
37
85
 5
5
84
16
63
 31
31
514
90
413
 4
4
73
18
94
 10
10
170
6
147
 16
16
260
13
324
 0.8
0.8
13
17
12
 5
5
78
27
100
 12
12
199
12
136
 5
5
84
43
130
 24
24
397
102
297
 16
16
262
12
298
 8
8
134
14
124
 7
7
121
8
92
 8
8
134
12
96
 9
9
144
12
95
 23
23
388
60
360
 6
6
101
21
116
 7
7
119
12
82
 9
9
156
12
104
 10
10
160
14
55
 27
27
451
18
305
 100
100
1659
43
5553
 8
8
128
76
371
 37
37
622
78
546
 4
4
65
48
60
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
99.8
100 99.7
99.7
99.8
98.5 -
-
-
95.5 -
-
-
- 35.8
35.8
44.8
94 51
51
63.5
- -
-
-
93 92.8
92.8
96.7
93.5 -
-
-
- 80.9
80.9
87.2
- 80.9
80.9
87.2
85 85.2
85.2
92.2
75.5 74.8
74.8
83.5
68 63.2
63.2
73.7
- -
-
-
45 20.4
20.4
30.1
- 40.3
40.3
52.7
- 20.7
20.7
29.6
- 38
38
52
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 28.1
28.1
42.4
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC2 - P06493 |
| 2 | LIMK1 - P53667 |
| 3 | ZYX - Q15942 |
| 4 | STK3 - Q13188 |
| 5 | STK4 - Q13043 |
| 6 | SAV1 - Q9H4B6 |
Regulation
Activation:
Activated by phosphorylation at Ser-909 and Thr-1079.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| NuaK1 | O60285 | S464 | NIPVRSNSFNNPLGN | - |
| NuaK2 | Q9H093 | S464 | NIPVRSNSFNNPLGN | - |
| CDK1 | P06493 | T490 | ATTVTAITPAPIQQP | |
| CDK1 | P06493 | S613 | EKKQITTSPITVRKN | |
| LATS1 | O95835 | S674 | EMMRVGLSQDAQDQM | |
| MST2 | Q13188 | S872 | GDHPRQDSMDFSNEW | + |
| MST2 | Q13188 | S909 | HQRCLAHSLVGTPNY | + |
| MST2 | Q13188 | T967 | MKVINWQTSLHIPPQ | |
| MST2 | Q13188 | T1012 | KAHPFFKTIDFSSDL | |
| LATS1 | O95835 | S1049 | VDPDKLWSDDNEEEN | |
| MST2 | Q13188 | T1060 | EEENVNDTLNGWYKN | |
| MST2 | Q13188 | T1079 | EHAFYEFTFRRFFDD | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| LATS1 | O95835 | S1049 | VDPDKLWSDDNEEEN | |
| LATS1 | O95835 | S674 | EMMRVGLSQDAQDQM | |
| MYPT1 (PPP1R12A) | O14974 | S445 | LGLRKTGSYGALAEI | + |
| TAZ | Q16635 | S66 | GSHSRQSSTDSSGGH | |
| TAZ | Q16635 | S89 | AQHVRSHSSPASLQL | - |
| YAP1 | P46937 | S109 | KSHSRQASTDAGTAG | |
| YAP1 | P46937 | S127 | PQHVRAHSSPASLQL | - |
| YAP1 | P46937 | S164 | AQHLRQSSFEIPDDV | |
| YAP1 | P46937 | S381 | LRTMTTNSSDPFLNS | |
| YAP1 | P46937 | S61 | IVHVRGDSETDLEAL |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
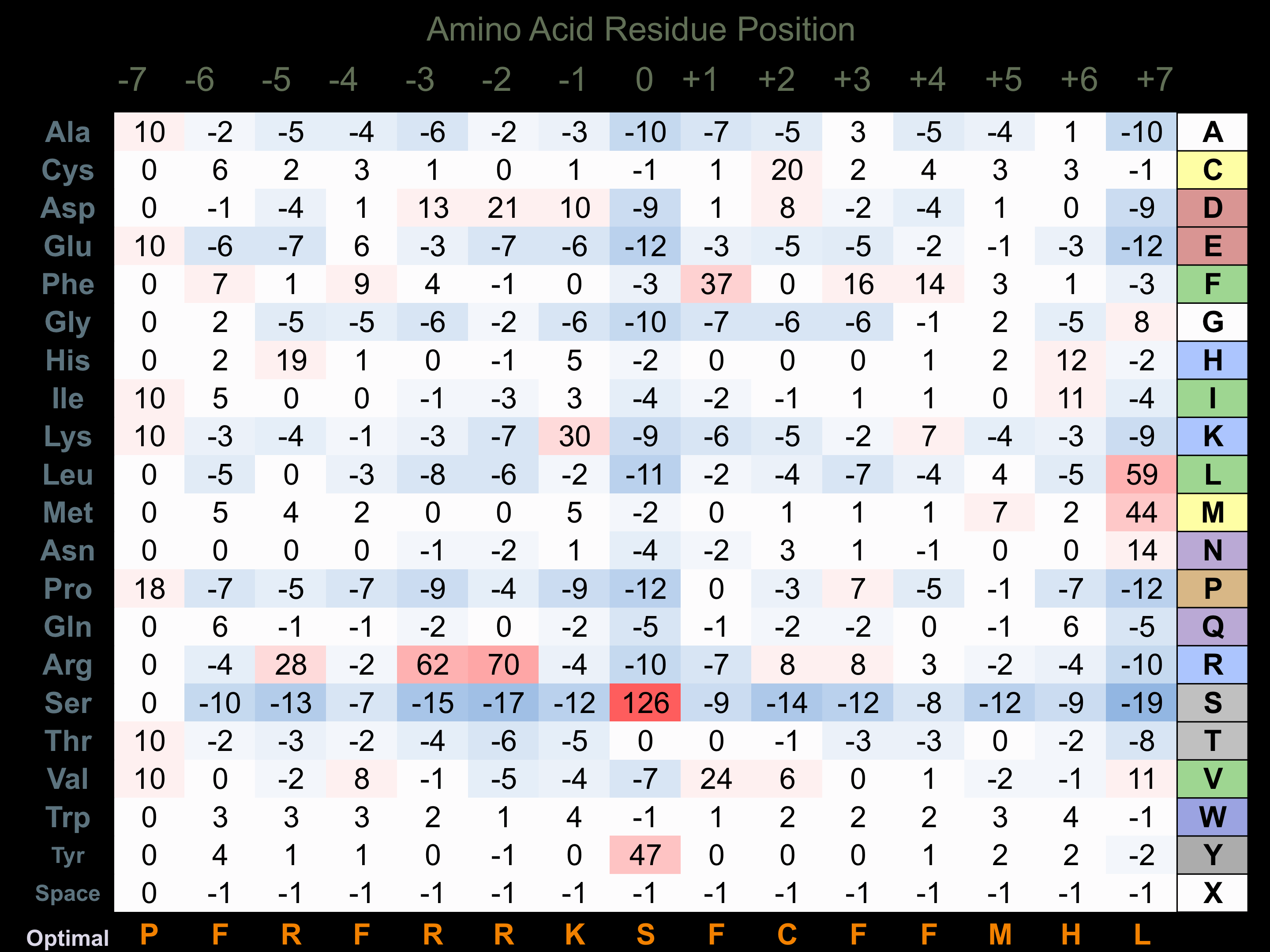
Matrix Type:
Experimentally derived from alignment of 8 known protein substrate phosphosites and 49 peptides phosphorylated by recombinant LATS1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Lestaurtinib | Kd = 30 nM | 126565 | 22037378 | |
| Staurosporine | Kd = 42 nM | 5279 | 18183025 | |
| Nintedanib | Kd = 420 nM | 9809715 | 502835 | 22037378 |
| AST-487 | Kd = 470 nM | 11409972 | 574738 | 18183025 |
| Sunitinib | Kd = 630 nM | 5329102 | 535 | 18183025 |
| SU14813 | Kd = 850 nM | 10138259 | 1721885 | 18183025 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| A674563 | Kd = 1.1 µM | 11314340 | 379218 | 22037378 |
| N-Benzoylstaurosporine | Kd = 1.1 µM | 56603681 | 608533 | 18183025 |
| NVP-TAE684 | Kd = 1.6 µM | 16038120 | 509032 | 22037378 |
| Dovitinib | Kd = 1.7 µM | 57336746 | 18183025 | |
| Linifanib | Kd = 2.1 µM | 11485656 | 223360 | 22037378 |
| Ruboxistaurin | Kd = 2.6 µM | 153999 | 91829 | 18183025 |
| JNJ-28312141 | Kd = 3.3 µM | 22037378 | ||
| TG100115 | Kd = 4.5 µM | 10427712 | 230011 | 22037378 |
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Tetraploidy; Mesoblastic nephroma; Childhood kidney neoplasm
Comments:
LATS1 appears to be a tumour suppressor protein (TSP), although it seems to undergo a typical rate bystander mutations in human cancers. The active form of the protein kinase normally acts to inhibit tumour cell proliferation. It also promotes apoptosis and inhibit the oncogene action of YAP1. Tetraploidy has a relation to the leukemia and myeloid leukemia disorders. Tetraploidy is a rare disease characterized by having 4 chromosomes (twice the usual number), leading to symptoms that could include radial club hand, beaked nose, cleft palate, and bifid uvula. Affected tissues can include myeloid, colon, and lung. Mesoblastic Nephroma is a kidney tumour made of expanded connective tissue cells (fibroblasts) and has the possibility of metastasizing. The rare condition of Childhood Kidney Neoplasm has a relation to clear cell sarcoman and Wilms tumour. NUAK1 and NUAK2 mediated phosphorylation of LATS1 can be inhibited with a LATS1 mutation of S464A. Inhibition of LATS1-YAP1 interaction can occur with a Y559F mutation. The K734A mutation can abrogate kinase phosphotransferase activity, lack of p53 translation, increased time for mitosis, autophosphorylation, and increased ploidy.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24888 diverse cancer specimens. This rate is the same as the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R82* (4); R82Q (2).
Comments:
Only 4 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.