Nomenclature
Short Name:
MST2
Full Name:
Serine-threonine-protein kinase 3
Alias:
- EC 2.7.11.1
- FLJ90748
- CLIK1
- Krs-1
- MESS1
- Serine,threonine protein kinase 3
- STK3
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE20
SubFamily:
MST
Structure
Mol. Mass (Da):
56,301
# Amino Acids:
491
# mRNA Isoforms:
2
mRNA Isoforms:
59,462 Da (519 AA; Q13188-2); 56,301 Da (491 AA; Q13188)
4D Structure:
Homodimer; mediated via the coiled-coil region. Interacts with NORE1, which inhibits autoactivation By similarity. Interacts with and stabilizes SAV1. Interacts with RAF1, which prevents dimerization and phosphorylation. Interacts with RASSF1, which leads to enzyme activation
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 27 | 278 | Pkinase |
| 287 | 323 | Coiled-coil |
| 437 | 484 | SARAH |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S15, S18, S316, S323, S333, S385, S392, S444..
Threonine phosphorylated:
T117-, T172+, T174+, T180+, T235, T240, T325, T336, T384-.
Tyrosine phosphorylated:
Y81, Y396.
Ubiquitinated:
K56, K116, K148, K161, K177, K243, K298.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 72
72
834
29
1008
 4
4
45
15
44
 5
5
59
2
74
 29
29
331
85
588
 61
61
700
24
645
 11
11
122
88
191
 16
16
188
31
416
 100
100
1155
33
2268
 33
33
383
17
316
 5
5
63
72
58
 4
4
43
19
45
 44
44
505
173
569
 2
2
18
24
19
 9
9
104
12
109
 3
3
34
14
27
 5
5
57
15
48
 22
22
249
112
1315
 2
2
25
10
16
 4
4
47
77
58
 48
48
552
109
649
 4
4
41
13
22
 3
3
33
16
30
 2
2
20
20
26
 3
3
34
11
24
 1.3
1.3
15
13
17
 80
80
922
51
1405
 2
2
18
27
16
 2
2
19
11
16
 6
6
66
11
46
 0.9
0.9
10
28
9
 46
46
535
24
541
 33
33
376
36
393
 18
18
205
67
418
 59
59
684
62
644
 16
16
189
35
287
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 86.8
86.8
86.8
100 78.6
78.6
89.8
100 -
-
-
100 -
-
-
96 99.4
99.4
99.6
99 -
-
-
- 95.8
95.8
97.2
97 96.7
96.7
98.4
97 -
-
-
- 53.2
53.2
56.4
- 79.4
79.4
88.2
98 93.7
93.7
96.3
94 89.4
89.4
93.5
89 -
-
-
- 46.2
46.2
58.6
46 66.4
66.4
75.6
- 51.5
51.5
67
74 65
65
78.6
- -
-
-
- -
-
-
- -
-
-
43 -
-
-
43 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | SAV1 - Q9H4B6 |
| 2 | CASP3 - P42574 |
| 3 | RAF1 - P04049 |
| 4 | LATS1 - O95835 |
Regulation
Activation:
Activated by caspase-cleavage. Full activation also requires homodimerization and autophosphorylation of Thr-180, which are inhibited by Raf1.
Inhibition:
Inhibited by the C-terminal non-catalytic region.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Abl1 | P00519 | T735 | DTEWRSVTLPRDLQS | + |
| LATS1 | O95835 | S872 | GDHPRQDSMDFSNEW | + |
| LATS1 | O95835 | S909 | HQRCLAHSLVGTPNY | + |
| LATS1 | O95835 | T1012 | KAHPFFKTIDFSSDL | |
| LATS1 | O95835 | T1060 | EEENVNDTLNGWYKN | |
| LATS1 | O95835 | T1079 | EHAFYEFTFRRFFDD | + |
| LATS1 | O95835 | T967 | MKVINWQTSLHIPPQ | |
| MOBKL1A (MOB1B) | Q7L9L4 | T12 | FGSRSSKTFKPKKNI | + |
| MOBKL1A (MOB1B) | Q7L9L4 | T35 | LLKHAEATLGSGNLR | + |
| MOBKL1B (MOB1A) | Q9H8S9 | T74 | QINMLYGTITEFCTE | + |
| MST2 (CLIK1, STK3) | Q13188 | T180 | DTMAKRNTVIGTPFW | + |
| YAP1 | P46937 | S127 | PQHVRAHSSPASLQL |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
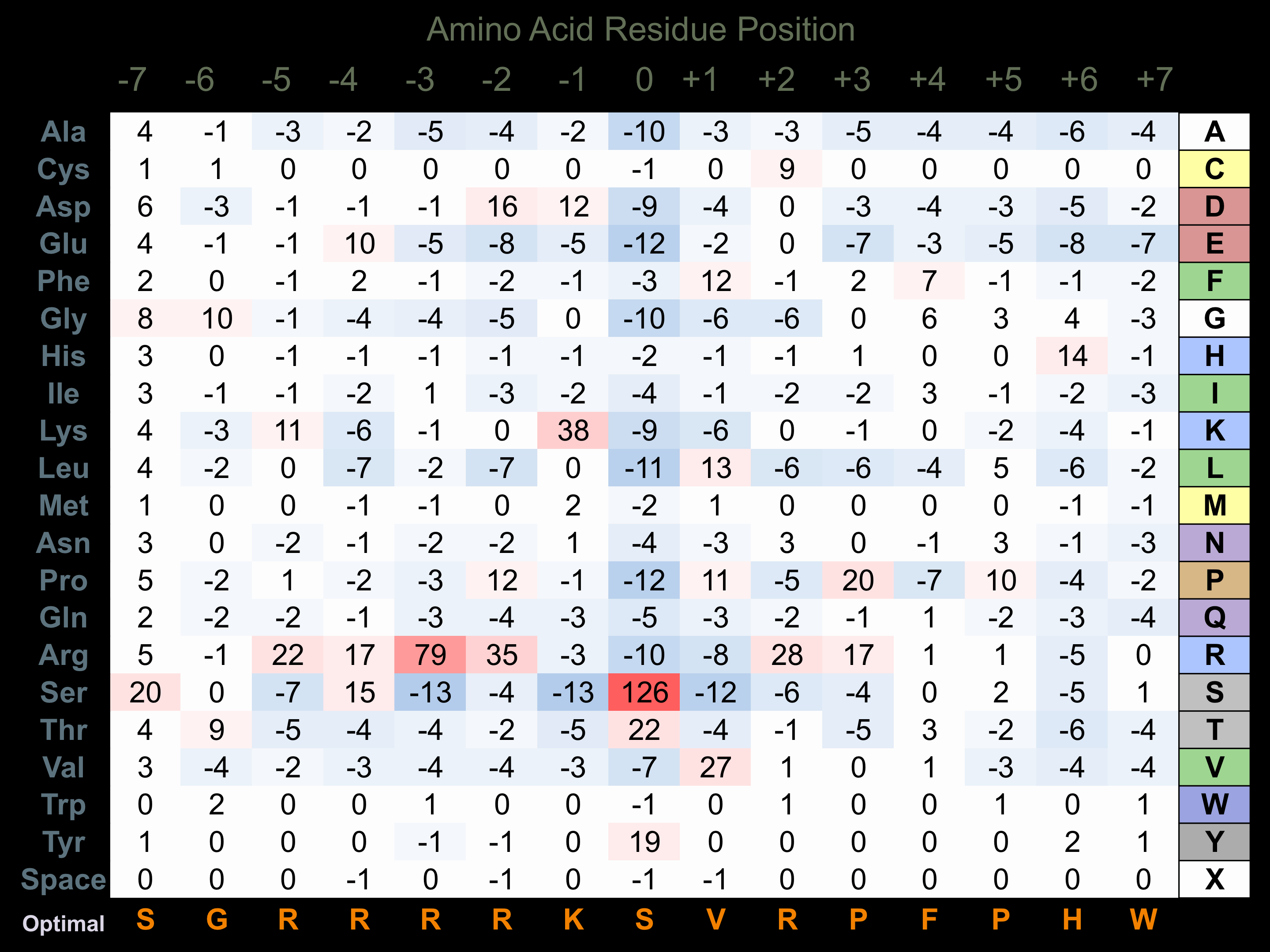
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Comments:
MST2 might be a tumour suppressor protein (TSP). MST2 may function in a way similar to a growth suppressor. The protein is critical in organ size control and tumour suppression by restricting proliferation and promoting apoptosis. MST2 levels are up-regulated almost 3-fold higher in human cancers when compared to most other protein kinases.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer (%CFC= -67, p<(0.0003); Colon mucosal cell adenomas (%CFC= +58, p<0.012); Head and neck squamous cell carcinomas (HNSCC) (%CFC= +68, p<0.005); and Oral squamous cell carcinomas (OSCC) (%CFC= +128, p<0.007). The COSMIC website notes an up-regulated expression score for MST2 in diverse human cancers of 1337, which is 2.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 76 for this protein kinase in human cancers was 1.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A K56R mutation can abrogate phosphotransferase activity, and D322N substitution can produce resistant to proteolytic cleavage.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25371 diverse cancer specimens. This rate is only -6 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.41 % in 602 endometrium cancers tested; 0.28 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,655 cancer specimens
Comments:
Only 3 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.

