Nomenclature
Short Name:
MEKK3
Full Name:
Mitogen-activated protein kinase kinase kinase 3
Alias:
- EC 2.7.11.25
- Kinase MEKK3
- MAPKKK3
- MEK kinase 3
- MEKK 3
- M3K3
- MAP/ERK kinase kinase 3
- MAP3K3
- MAPK/ERK kinase kinase 3
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE11
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
70898
# Amino Acids:
626
# mRNA Isoforms:
2
mRNA Isoforms:
74,089 Da (657 AA; Q99759-2); 70,898 Da (626 AA; Q99759)
4D Structure:
Binds both upstream activators and downstream substrates in multimolecular complexes. Part of a complex with MAP2K3, RAC1 and CCM2. Interacts with MAP2K5 and SPAG9
1D Structure:
Subfamily Alignment

Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S130, S131, S145, S147, S162, S166-, S168, S169, S175, S176, S194, S234, S237, S250, S289, S312, S314, S316, S337-, S340, S345, S355, S357, S464, S520+, S526+.
Threonine phosphorylated:
T274, T294, T317, T516+, T528+, T530-.
Tyrosine phosphorylated:
Y155, Y275, Y288, Y427, Y428, Y445.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1387
28
1162
 7
7
104
10
56
 15
15
203
16
138
 18
18
247
117
505
 59
59
812
33
636
 3
3
44
55
27
 14
14
190
39
477
 26
26
355
45
556
 24
24
332
10
270
 11
11
148
113
105
 8
8
105
35
95
 45
45
631
138
706
 12
12
162
27
104
 5
5
72
6
38
 5
5
73
32
51
 6
6
79
17
65
 17
17
241
248
2089
 11
11
153
21
132
 8
8
112
94
84
 50
50
690
112
596
 10
10
140
31
146
 12
12
169
33
143
 9
9
129
25
134
 12
12
165
21
116
 10
10
139
31
118
 82
82
1139
76
2008
 10
10
138
30
102
 10
10
137
21
87
 14
14
195
21
154
 7
7
99
42
93
 61
61
842
18
550
 64
64
893
31
1380
 9
9
126
117
240
 57
57
796
83
724
 10
10
143
57
133
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 66
66
78.6
100 89.5
89.5
90.9
100 -
-
-
97 -
-
-
99 97.6
97.6
99
98 -
-
-
- 96.7
96.7
98.6
97 96.3
96.3
98.4
96 -
-
-
- 60.2
60.2
65.5
- 85.6
85.6
91
88 61
61
74.6
- 77.8
77.8
85.9
79 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 39.3
39.3
48.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 26.2
26.2
41.3
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | YWHAE - P62258 |
| 2 | HSPA2 - P54652 |
| 3 | TRAF7 - Q6Q0C0 |
| 4 | RCAN1 - P53805 |
| 5 | GAB1 - Q13480 |
| 6 | NFKBIA - P25963 |
| 7 | PRKACA - P17612 |
| 8 | WNK1 - Q9H4A3 |
| 9 | HSPA4 - P34932 |
| 10 | PAK1 - Q13153 |
Regulation
Activation:
Activated by phosphorylation on Thr-530
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ANKRD3 (RIPK4) | P57078 | T184 | SMDGLFGTIAYLPPE | + |
| DSCR1 | P53805 | S163 | PDKQFLISPPASPPV | |
| DSCR1 | P53805 | S167 | FLISPPASPPVGWKQ | |
| MKK3 (MAP2K3, MEK3) | P46734 | S218 | ISGYLVDSVAKTMDA | + |
| MKK3 (MAP2K3, MEK3) | P46734 | T222 | LVDSVAKTMDAGCKP | + |
| MKK6 (MAP2K6, MEK6) | P52564 | S207 | ISGYLVDSVAKTIDA | + |
| MKK6 (MAP2K6, MEK6) | P52564 | T211 | LVDSVAKTIDAGCKP | + |
| MKK7 (MAP2K7, MEK7) | O14733 | S271 | ISGRLVDSKAKTRSA | + |
| MKK7 (MAP2K7, MEK7) | O14733 | T275 | LVDSKAKTRSAGCAA | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
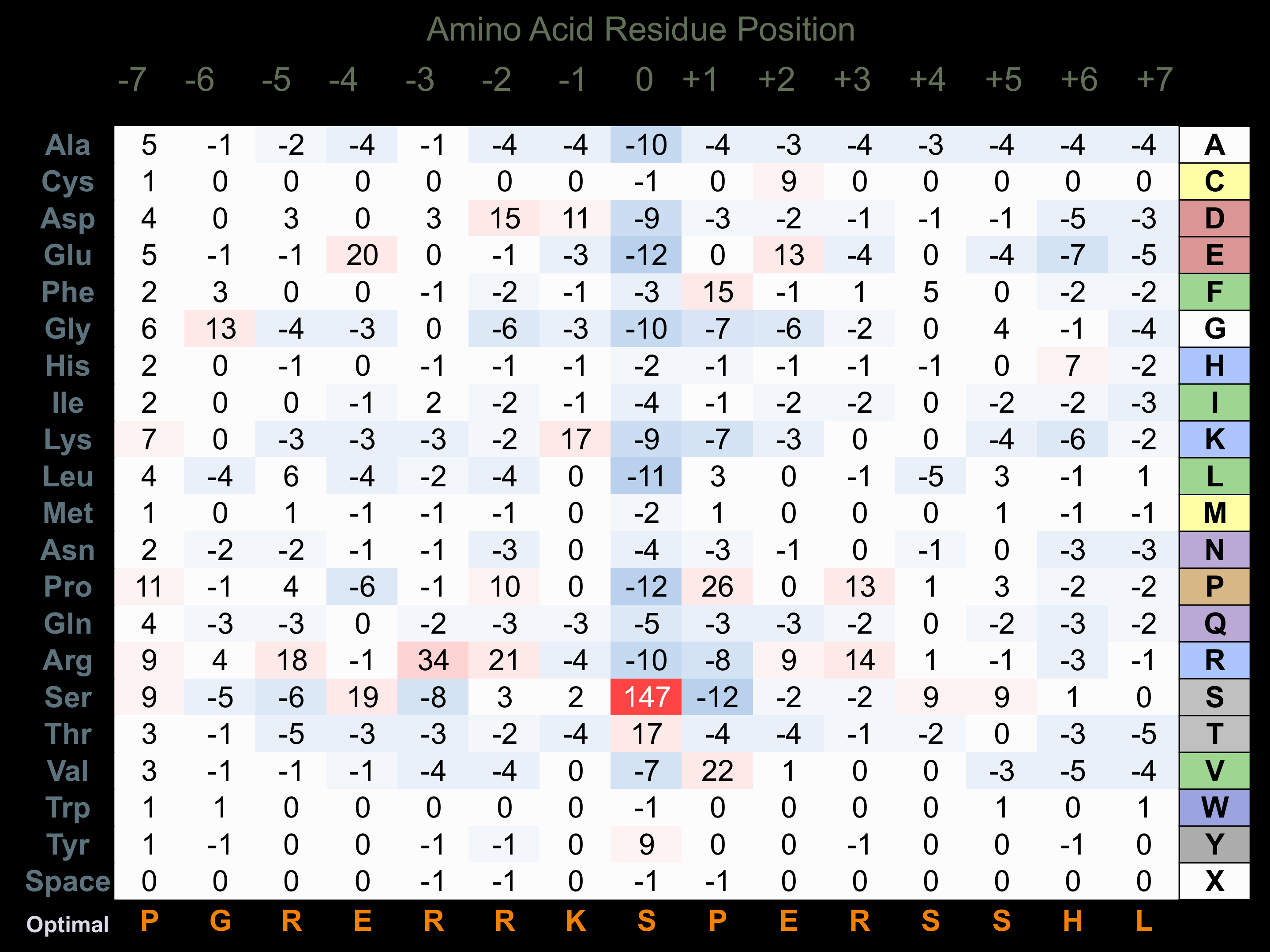
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Esophageal cancer
Comments:
MEKK3 may be an oncoprotein (OP). Significantly elevated expression of MEKK3 was observed in esophageal squamous cell carcinoma (ESCC) in the early stages of tumorigenesis, indicating an important role for the protein in the development of the cancer. In addition, patients with MEKK3-positive ESCC tumours displayed signficantly reduced mean survival time (~9 months) compared to patients with MEKK3-negative tumours (>21 months). Therefore, elevated MEKK3 expression is suggested to play an important role in the early tumorigenesis of ESCC.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Colorectal adenocarcinomas (early onset) (%CFC= +82, p<0.005); Skin fibrosarcomas (%CFC= +84); Skin melanomas - malignant (%CFC= +80, p<0.0001); and Skin squamous cell carcinomas (%CFC= -46, p<0.049). The COSMIC website notes an up-regulated expression score for MEKK3 in diverse human cancers of 383, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 73 for this protein kinase in human cancers was 1.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25389 diverse cancer specimens. This rate is only -4 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.39 % in 1093 large intestine cancers tested; 0.32 % in 805 skin cancers tested; 0.27 % in 602 endometrium cancers tested; 0.27 % in 589 stomach cancers tested; 0.13 % in 1942 lung cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,655 cancer specimens
Comments:
Only 2 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.

