Nomenclature
Short Name:
MEK3
Full Name:
Dual specificity mitogen-activated protein kinase kinase 3
Alias:
- EC 2.7.12.2
- MAP kinase kinase 3
- MP2K3
- PRKMK3
- MAPK,ERK kinase 3
- MAPK/ERK kinase 3
- MAPKK 3
- MAP2K3
Classification
Type:
Dual specificity protein kinase
Group:
STE
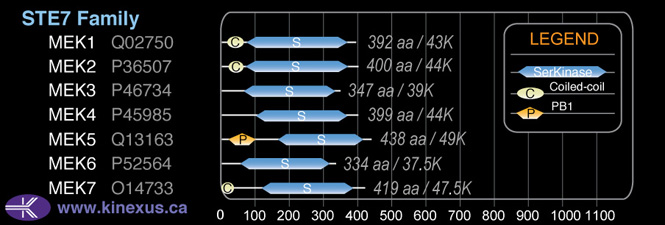
Family:
STE7
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
39,318
# Amino Acids:
347
# mRNA Isoforms:
3
mRNA Isoforms:
39,940 Da (352 AA; P46734-3); 39,318 Da (347 AA; P46734); 36,173 Da (318 AA; P46734-2)
4D Structure:
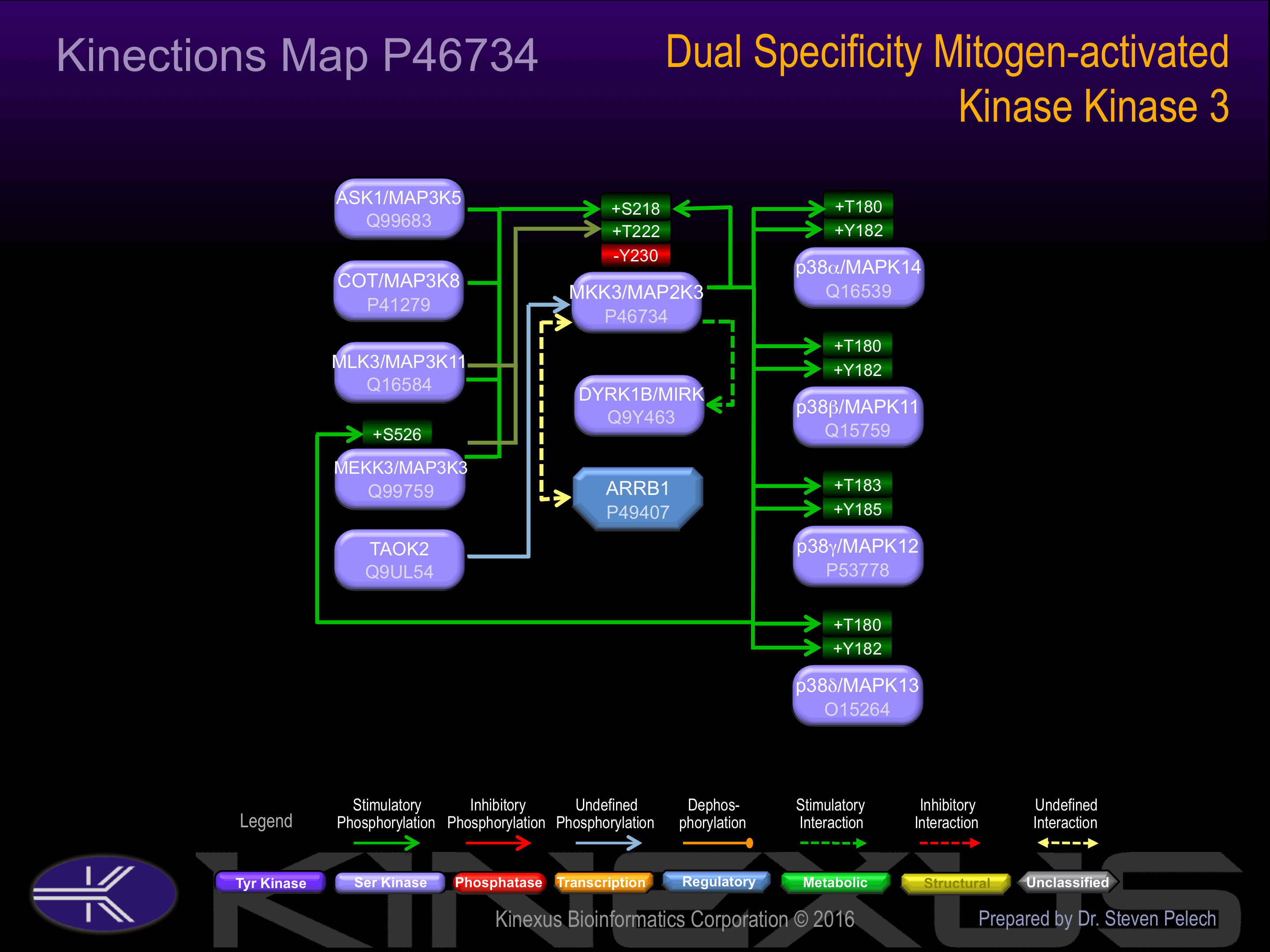
Binds to DYRK1B/MIRK and increases its kinase activity. Part of a complex with MAP3K3, RAC1 and CCM2. Interacts with ARRB1. Interacts with Yersinia yopJ.
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 64 | 325 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S3, S15, S28, S31, S101, S212, S218+, S249, S253.
Threonine phosphorylated:
T39, T222+.
Tyrosine phosphorylated:
Y214, Y230-, Y245.
Ubiquitinated:
K80, K183, K192, K243, K279, K340.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 9
9
1130
42
1114
 0.9
0.9
104
28
85
 2
2
270
16
620
 4
4
498
143
916
 8
8
914
42
753
 1.2
1.2
143
125
144
 2
2
296
53
513
 7
7
887
68
1246
 5
5
592
30
539
 2
2
216
134
166
 2
2
263
51
227
 7
7
861
263
739
 1.1
1.1
137
49
73
 0.5
0.5
65
24
55
 1.2
1.2
147
42
262
 1.4
1.4
168
23
118
 6
6
689
229
3761
 1.4
1.4
174
34
272
 4
4
429
139
266
 6
6
725
168
686
 0.7
0.7
81
39
93
 2
2
193
45
184
 2
2
202
27
163
 0.8
0.8
96
35
247
 1.2
1.2
149
39
199
 10
10
1181
91
1284
 2
2
190
58
149
 2
2
197
35
255
 0.7
0.7
87
35
84
 100
100
12102
42
6032
 4
4
481
42
280
 7
7
890
45
587
 3
3
406
88
420
 9
9
1029
83
809
 2
2
183
66
389
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 30.8
30.8
47.1
100 -
-
-
- -
-
-
96.5 -
-
-
- 99.1
99.1
99.7
99 -
-
-
- 96.5
96.5
98.8
96.5 37.2
37.2
56.8
96.5 -
-
-
- 89.8
89.8
93.2
- 30.4
30.4
48.7
91 42.1
42.1
58.1
- 75.3
75.3
84.5
- -
-
-
- 57.1
57.1
72.9
63 -
-
-
- 37.7
37.7
55.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 33.7
33.7
52.3
- 23.6
23.6
35.6
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAPK14 - Q16539 |
| 2 | TAOK2 - Q9UL54 |
| 3 | ELK1 - P19419 |
| 4 | DCTN1 - Q14203 |
| 5 | TAOK1 - Q7L7X3 |
| 6 | MAPK12 - P53778 |
| 7 | SMAD7 - O15105 |
| 8 | DYRK1B - Q9Y463 |
| 9 | MAP2K6 - P52564 |
| 10 | NPHS1 - O60500 |
| 11 | PRODH2 - Q9UF12 |
| 12 | PKN1 - Q16512 |
| 13 | MAPK8IP2 - Q13387 |
| 14 | MAPK3 - P27361 |
| 15 | PLCB2 - Q00722 |
Regulation
Activation:
Phosphorylation of Ser-218 and Thr-222 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| MEKK3 (MAP3K3) | Q99759 | S526 | MSGTGMRSVTGTPYW | + |
| MKK3 (MAP2K3, MEK3) | P46734 | S218 | ISGYLVDSVAKTMDA | + |
| p38a MAPK (MAPK14) | Q16539 | T180 | RHTDDEMTGYVATRW | + |
| p38a MAPK (MAPK14) | Q16539 | Y182 | TDDEMTGYVATRWYR | + |
Protein Kinase Specificity
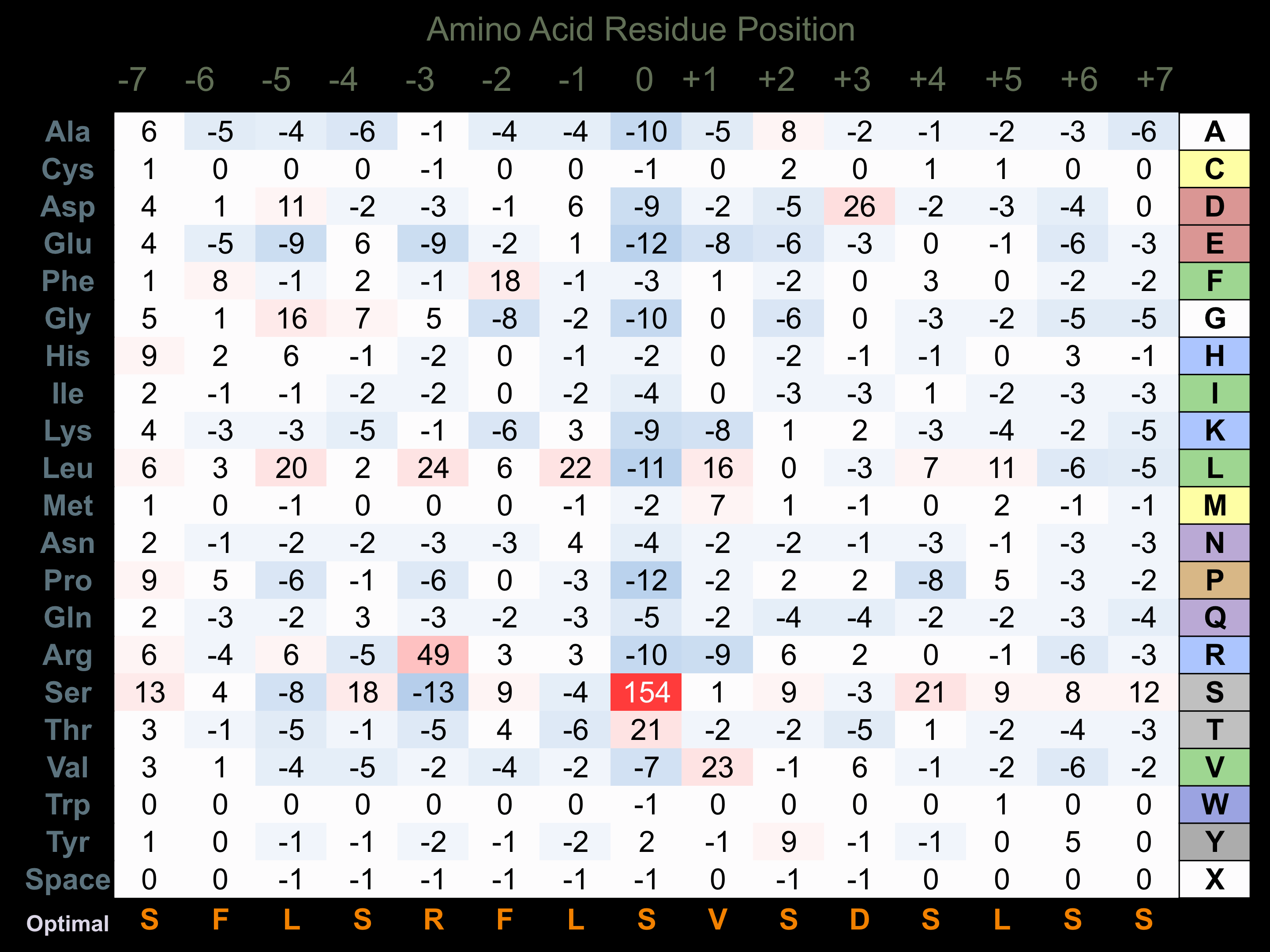
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Lestaurtinib | Kd = 5 nM | 126565 | 22037378 | |
| Staurosporine | Kd = 5 nM | 5279 | 18183025 | |
| PP242 | Kd = 92 nM | 25243800 | 22037378 | |
| R406 | Kd = 100 nM | 11984591 | 22037378 | |
| KW2449 | Kd = 150 nM | 11427553 | 1908397 | 22037378 |
| NVP-TAE684 | Kd = 170 nM | 16038120 | 509032 | 22037378 |
| JNJ-28312141 | Kd = 360 nM | 22037378 | ||
| Ruxolitinib | Kd = 470 nM | 25126798 | 1789941 | 22037378 |
| GSK1838705A | Kd = 1 µM | 25182616 | 464552 | 22037378 |
| MEK Inhibitor I | IC50 > 1 µM | 9951490 | 37493 | 15006386 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| A674563 | Kd = 1.2 µM | 11314340 | 379218 | 22037378 |
| Alvocidib | Kd = 1.2 µM | 9910986 | 428690 | 22037378 |
| JNJ-7706621 | Kd = 1.5 µM | 5330790 | 191003 | 18183025 |
| Sunitinib | Kd = 1.7 µM | 5329102 | 535 | 22037378 |
| Nintedanib | Kd = 2.1 µM | 9809715 | 502835 | 22037378 |
| AST-487 | Kd = 2.2 µM | 11409972 | 574738 | 22037378 |
| SU14813 | Kd = 2.2 µM | 10138259 | 1721885 | 22037378 |
| Bosutinib | Kd = 2.6 µM | 5328940 | 288441 | 22037378 |
| BMS-690514 | Kd < 3 µM | 11349170 | 21531814 | |
| Crizotinib | Kd = 3.3 µM | 11626560 | 601719 | 22037378 |
| TG101348 | Kd = 3.5 µM | 16722836 | 1287853 | 22037378 |
Disease Linkage
General Disease Association:
Cancer
Comments:
The protein regulates mitochondrial biogenesis in sepsis-induced lung injury.
Specific Cancer Types:
Colon cancer
Comments:
Defects in the MEK3 gene may be involved colon cancer. S218A and T222A substitutions are involved in inactivation. S218E and T222E mutations are associated in constitutive activation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Breast epithelial carcinomas (%CFC= +100, p<0.063); Ovary adenocarcinomas (%CFC= +141, p<0.019); and Uterine leiomyomas from fibroids (%CFC= -45, p<0.021). The COSMIC website notes an up-regulated expression score for MEK3 in diverse human cancers of 279, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 126 for this protein kinase in human cancers was 2.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.11 % in 25183 diverse cancer specimens. This rate is a modest 1.53-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.72 % in 805 skin cancers tested; 0.53 % in 1093 large intestine cancers tested; 0.3 % in 1753 lung cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,466 cancer specimens
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.