Nomenclature
Short Name:
MLK3
Full Name:
Mitogen-activated protein kinase kinase kinase 11
Alias:
- EC 2.7.11.25
- Kinase MLK3
- Mitogen activated protein kinase kinase kinase 11
- Mixed lineage kinase 3
- MLK-3
- SPRK
- M3K11
- MAP3K11
- MAPKKK11
- MEKK11
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
MLK
SubFamily:
MLK
Specific Links
Structure
Mol. Mass (Da):
92,688
# Amino Acids:
847
# mRNA Isoforms:
2
mRNA Isoforms:
92,688 Da (847 AA; Q16584); 65,344 Da (590 AA; Q16584-2)
4D Structure:
Homodimer; undergoes dimerization during activation.
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 41 | 105 | SH3 |
| 117 | 376 | TyrKc |
| 396 | 441 | Coiled-coil |
| 403 | 424 | Leucine zipper 1 |
| 438 | 459 | Leucine zipper 2 |
| 117 | 384 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
R512.
Serine phosphorylated:
S11, S35, S281+, S394, S493, S507, S524, S548, S555, S556, S654, S674, S693, S705, S724, S727, S740, S746, S748, S758, S770, S789, S793, S815.
Threonine phosphorylated:
T277+, T477, T526, T677, T702, T708, T738, T752, T755.
Ubiquitinated:
K264.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 62
62
667
28
762
 15
15
159
13
164
 10
10
111
11
95
 25
25
263
81
456
 70
70
750
21
684
 12
12
133
52
184
 41
41
433
33
668
 61
61
651
34
1367
 67
67
720
10
614
 45
45
478
66
583
 19
19
207
28
166
 61
61
653
143
647
 17
17
178
22
113
 12
12
129
11
110
 12
12
133
25
110
 10
10
102
14
94
 21
21
223
199
1074
 9
9
101
20
77
 11
11
115
59
79
 57
57
606
79
549
 17
17
185
23
201
 33
33
351
26
509
 10
10
107
20
76
 10
10
110
18
134
 19
19
207
23
210
 86
86
914
54
1506
 12
12
126
25
75
 11
11
121
19
101
 11
11
121
19
108
 30
30
326
14
93
 100
100
1069
18
656
 49
49
522
26
663
 5
5
54
69
101
 69
69
735
57
638
 9
9
101
35
51
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.4
99.4
99.6
99 98.3
98.3
98.8
98 -
-
-
94.5 -
-
-
100 82.6
82.6
83.7
93 -
-
-
- 94.4
94.4
95.4
95 93.8
93.8
95.4
94 -
-
-
- -
-
-
- -
-
-
- 43.3
43.3
56.6
- 46.5
46.5
58.7
- -
-
-
- 30.3
30.3
43.8
- 35.6
35.6
49.2
- 20.6
20.6
35.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
36 -
-
-
36 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC42 - P60953 |
| 2 | SH3RF1 - Q7Z6J0 |
| 3 | CHUK - O15111 |
| 4 | MAP4K1 - Q92918 |
| 5 | DLG4 - P78352 |
| 6 | AKT1 - P31749 |
| 7 | MAP4K2 - Q12851 |
| 8 | IKBKB - O14920 |
| 9 | RHOG - P84095 |
| 10 | MAP2K4 - P45985 |
| 11 | MAP3K12 - Q12852 |
| 12 | CDC37 - Q16543 |
| 13 | HSP90AA1 - P07900 |
| 14 | HSP90AA2 - Q14568 |
| 15 | MAPK8IP1 - Q9UQF2 |
Regulation
Activation:
Homodimerization via the leucine zipper domains is required for autophosphorylation and subsequent activation. Phosphorylation at Thr-277 and Ser-281 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| MKK3 (MAP2K3, MEK3) | P46734 | S218 | ISGYLVDSVAKTMDA | + |
| MKK3 (MAP2K3, MEK3) | P46734 | T222 | LVDSVAKTMDAGCKP | + |
| MKK4 (MAP2K4, MEK4) | P45985 | S257 | ISGQLVDSIAKTRDA | + |
| MKK4 (MAP2K4, MEK4) | P45985 | T261 | LVDSIAKTRDAGCRP | + |
| MKK7 (MAP2K7, MEK7) | O14733 | S271 | ISGRLVDSKAKTRSA | + |
| MKK7 (MAP2K7, MEK7) | O14733 | T275 | LVDSKAKTRSAGCAA | + |
| MLK3 (MAP3K11) | Q16584 | S281 | WHKTTQMSAAGTYAW | + |
| MLK3 (MAP3K11) | Q16584 | T277 | LAREWHKTTQMSAAG | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
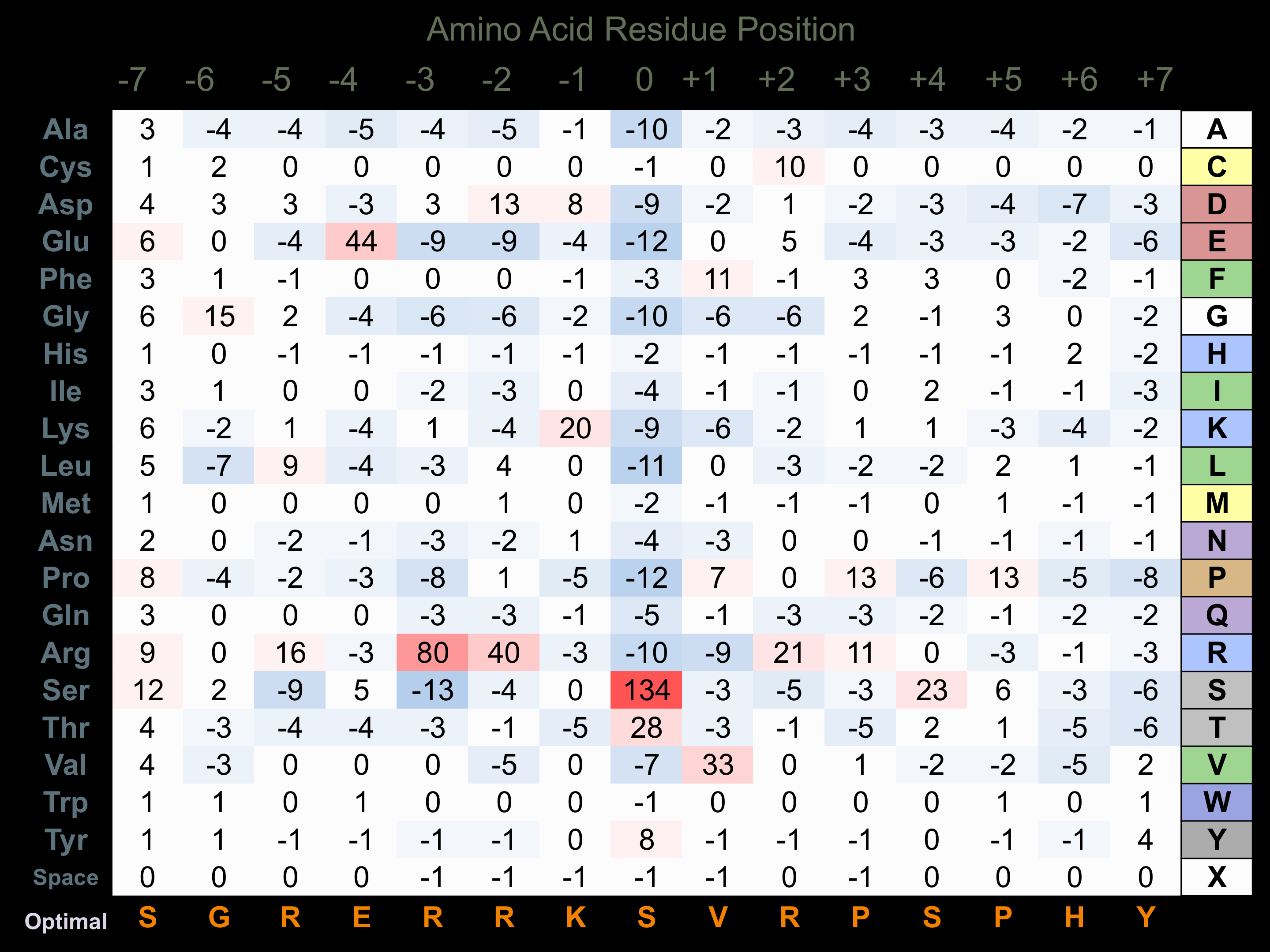
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Neurological and liver disorders
Specific Diseases (Non-cancerous):
HIV-1-associated dementia (HAD); Non-alcoholic steatohepatitis (NASH)
Comments:
The HIV-1 encoded proteins Tat and gp120 are toxic to neurons and activate monocytes and other myeloid cells, which may contribute the clinical manifestation of Human immunodeficiency virus (HIV)-1-associated dementia (HAD) and other HIV-1-associated neurocognitive disorders (HAND). Greater than 50% of patients living with HIV-1 will develop HAND, even with stringent adherence to anti-retroviral therapy. Since over -expression of MLK3 in rat primary rat neurons induces their apoptosis, and selective inhibition of MLK3 protects Tat and gp120-mediated activation of monocytes (with the release of inflammatory cytokines), MLK3 may be a useful target for treatment of HAD and HAND. MLK3 has been implicated as a regulator of disease progression in Non-alcoholic steatohepatitis (NASH), which is one of the most common chronic liver diseases worldwide.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +118, p<0.006); Brain glioblastomas (%CFC= +743, p<0.002); Brain oligodendrogliomas (%CFC= +1685, p<0.009); Large B-cell lymphomas (%CFC= +49, p<0.039); Ovary adenocarcinomas (%CFC= +127, p<0.031); Pituitary adenomas (ACTH-secreting) (%CFC= -49); Skin melanomas - malignant (%CFC= +59, p<0.0001); Skin squamous cell carcinomas (%CFC= +47, p<0.026); and T-cell prolymphocytic leukemia (%CFC= +71, p<0.009). The COSMIC website notes an up-regulated expression score for MLK3 in diverse human cancers of 439, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 114 for this protein kinase in human cancers was 1.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25371 diverse cancer specimens. This rate is only -11 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.43 % in 1093 large intestine cancers tested; 0.37 % in 805 skin cancers tested; 0.24 % in 589 stomach cancers tested; 0.1 % in 1941 lung cancers tested; 0.09 % in 1270 liver cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,655 cancer specimens
Comments:
Only 6 deletions, 1 insertion and no complex mutations are noted on the COSMIC website.

