Nomenclature
Short Name:
Akt1
Full Name:
RAC-alpha serine-threonine-protein kinase
Alias:
- AKT
- Kinase Akt1
- PKB
- PKB-alpha
- PRKBA
- V-akt murine thymoma viral oncogene 1; Protein kinase B; RAC; RAC-alpha serine,threonine kinase; RAC-alpha serine/threonine kinase; RAC-PK-alpha
- Akt1
- AKT1 kinase
- C-AKT
- EC 2.7.11.1
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
AKT
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
55,686
# Amino Acids:
480
# mRNA Isoforms:
2
mRNA Isoforms:
55,686 Da (480 AA; P31749); 48,347 Da (418 AA; P31749-2)
4D Structure:
Interacts with AGAP2 isoform 2 (PIKE-A) in the presence of guanine nucleotides. The C-terminus interacts with CCDC88A/GRDN and THEM4. Interacts with AKTIP. Interacts (via PH domain) with MTCP1, TCL1A AND TCL1B. Interacts with CDKN1B; the interaction phosphorylates CDKN1B promoting 14-3-3 binding and cell-cycle progression. Interacts with TRAF6. Interacts with GRB10; the interaction leads to GRB10 phosphorylation thus promoting YWHAE/14-3-3-binding
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
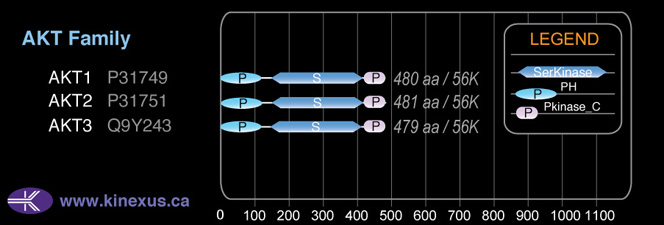
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K420, K426.
O-GlcNAcylated:
S126, S129, T305, T308, T312, S473.
Serine phosphorylated:
S122, S124+, S126, S129+, S137, S246+, S378, S396, S457, S473+, S475, S477.
Sumoylated:
K276, K301.
Threonine phosphorylated:
T34-, T65, T72+, T87, T92, T146, T211, T291, T305, T308+, T312-, T435, T443, T448, T450+, T479.
Tyrosine phosphorylated:
Y176+, Y315-, Y326+, Y417, Y437, Y474+.
Ubiquitinated:
K8, K14, K30, K276, K377.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 24
24
876
16
972
 8
8
305
10
188
 7
7
267
14
169
 27
27
1003
69
1448
 28
28
1015
13
890
 14
14
511
45
879
 12
12
430
19
610
 48
48
1768
39
3146
 16
16
571
10
505
 6
6
219
60
146
 8
8
309
29
204
 20
20
743
120
695
 6
6
229
25
176
 3
3
128
9
117
 8
8
288
26
243
 8
8
308
8
193
 19
19
684
117
3317
 5
5
193
21
148
 5
5
181
63
118
 19
19
700
56
708
 5
5
196
25
130
 9
9
329
27
233
 10
10
370
23
238
 3
3
108
21
96
 6
6
228
25
174
 69
69
2538
45
4715
 4
4
161
28
112
 8
8
280
21
245
 6
6
223
21
164
 6
6
218
14
116
 23
23
842
30
460
 100
100
3672
21
6348
 6
6
233
54
329
 25
25
906
31
727
 5
5
166
22
166
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 81.5
81.5
92.1
91 99.6
99.6
99.6
100 -
-
-
96 -
-
-
96 94
94
95.2
97 -
-
-
- 98.1
98.1
98.8
98 98.1
98.1
99.2
98 -
-
-
- 73
73
80.5
- 39.8
39.8
56.9
96 93.1
93.1
96.3
93 39.4
39.4
57.5
83 -
-
-
- 49.6
49.6
61.7
- -
-
-
- 52.5
52.5
67.8
60.5 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | GSK3B - P49841 |
| 2 | ILK - Q13418 |
| 3 | PDPK1 - O15530 |
| 4 | PDPK2 - Q6A1A2 |
| 5 | HSP90AA1 - P07900 |
| 6 | MDM2 - Q00987 |
| 7 | HSP90AA2 - Q14568 |
| 8 | TERT - O14746 |
| 9 | TCL1A - P56279 |
| 10 | TSC2 - P49815 |
| 11 | YBX1 - P67809 |
| 12 | PAK1 - Q13153 |
| 13 | AR - P10275 |
| 14 | FOXO1 - Q12778 |
| 15 | BRCA1 - P38398 |
Regulation
Activation:
Phosphorylation of Thr-72, Ser-124, Ser-129, Ser-246, Thr-308, Tyr-315, Tyr-326, Thr-450, Ser-473 and Tyr-474 increases phosphotransferase activity. TRAF6-induced 'Lys-63'-linked AKT1 ubiquitination is critical for phosphorylation and activation.
Inhibition:
Phosphorylation of Thr-34 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
When fully phosphorylated and translocated into the nucleus, undergoes 'Lys-48'-polyubiquitination catalyzed by TTC3, leading to its degradation by the proteasome.
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKCz | Q05513 | T34 | FLLKNDGTFIGYKER | - |
| Akt1 | P31749 | T72 | TERPRPNTFIIRCLQ | + |
| CK2a1 | P68400 | S129 | SGSPSDNSGAEEMEV | + |
| ACK1 | Q07912 | Y176 | EKATGRYYAMKILKK | + |
| Akt1 | P31749 | S246 | LSRERVFSEDRARFY | + |
| ERK1 | P27361 | T308 | KDGATMKTFCGTPEY | + |
| PKCb | P05771 | T308 | KDGATMKTFCGTPEY | + |
| PKCe | Q02156 | T308 | KDGATMKTFCGTPEY | + |
| CaMKK1 | Q8N5S9 | T308 | KDGATMKTFCGTPEY | + |
| PKCa | P17252 | T308 | KDGATMKTFCGTPEY | + |
| PDK1 | O15530 | T308 | KDGATMKTFCGTPEY | + |
| PIK3CA | P42336 | T308 | KDGATMKTFCGTPEY | + |
| PKD1 | Q15139 | T308 | KDGATMKTFCGTPEY | + |
| p38a | Q16539 | T308 | KDGATMKTFCGTPEY | + |
| SRC | P12931 | Y315 | TFCGTPEYLAPEVLE | - |
| RET | P07949 | Y315 | TFCGTPEYLAPEVLE | - |
| SRC | P12931 | Y326 | EVLEDNDYGRAVDWW | + |
| BRK | Q13882 | Y326 | EVLEDNDYGRAVDWW | + |
| ATM | Q13315 | S473 | RPHFPQFSYSASGTA | + |
| ATR | Q13535 | S473 | RPHFPQFSYSASGTA | + |
| PKCb | P05771 | S473 | RPHFPQFSYSASGTA | + |
| PKCe | Q02156 | S473 | RPHFPQFSYSASGTA | + |
| MAPKAPK5 | Q8IW41 | S473 | RPHFPQFSYSASGTA | + |
| DNAPK | P78527 | S473 | RPHFPQFSYSASGTA | + |
| FRAP1 | P42345 | S473 | RPHFPQFSYSASGTA | + |
| PKCa | P17252 | S473 | RPHFPQFSYSASGTA | + |
| PDK1 | O15530 | S473 | RPHFPQFSYSASGTA | + |
| PIK3CA | P42336 | S473 | RPHFPQFSYSASGTA | + |
| ILK | Q13418 | S473 | RPHFPQFSYSASGTA | + |
| PKD1 | Q15139 | S473 | RPHFPQFSYSASGTA | + |
| p38a | Q16539 | S473 | RPHFPQFSYSASGTA | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 14-3-3 zeta (YWHAZ) | P63104 | S58 | VVGARRSSWRVVSSI | |
| Acinus | Q9UKV3 | S1180 | GPRSRSRSRDRRRKE | |
| ACLY | P53396 | S455 | PAPSRTASFSESRAD | |
| ADRB2 | P07550 | S346 | LLCLRRSSLKAYGNG | |
| Akt1 (PKBa) | P31749 | S246 | LSRERVFSEDRARFY | + |
| Akt1 (PKBa) | P31749 | T72 | TERPRPNTFIIRCLQ | + |
| Akt1S1 | Q96B36 | S221 | DLDRIAASMRALVLR | |
| Akt1S1 | Q96B36 | T246 | LPRPRLNTSDFQKLK | |
| AMPKa2 (PRKAA2) | P54646 | S491 | STPQRSCSAAGLHRP | |
| APS | O14492 | S598 | SARSRSNSAERLLEA | |
| AR | P10275 | S213 | SGRAREASGAPTSSK | - |
| AR | P10275 | S791 | CVRMRHLSQEFGWLQ | ? |
| Arfaptin 2 | P53365 | S260 | GTRGRLESAQATFQA | |
| AS160 (TBC1D4) | O60343 | S588 | RMRGRLGSVDSFERS | - |
| AS160 (TBC1D4) | O60343 | T642 | QFRRRAHTFSHPPSS | - |
| ASK1 (MAP3K5) | Q99683 | S83 | ATRGRGSSVGGGSRR | - |
| ataxin-1 | P54253 | S776 | ATRKRRWSAPESRKL | |
| B-Raf | P15056 | S365 | GQRDRSSSAPNVHIN | - |
| B-Raf | P15056 | S429 | PQRERKSSSSSEDRN | - |
| Bad | Q92934 | S118 | GRELRRMSDEFVDSF | - |
| Bad | Q92934 | S75 | EIRSRHSSYPAGTED | - |
| Bad | Q92934 | S99 | PFRGRSRSAPPNLWA | - |
| Baxa | Q07812 | S184 | VAGVLTASLTIWKKM | - |
| BRCA1 | P38398 | T509 | LKRKRRPTSGLHPED | |
| BRF1 | Q92994 | S92 | RFRDRSFSEGGERLL | |
| CaRHSP1 | Q9Y2V2 | S52 | TRRTRTFSATVRASQ | |
| Caspase 9 | P55211 | S196 | KLRRRFSSLHFMVEV | - |
| CCT2 | P78371 | S260 | GSRVRVDSTAKVAEI | |
| CDK2 | P24941 | T39 | LKKIRLDTETEGVPS | ? |
| CENTB1 | Q15027 | S554 | SIRPRPGSLRSKPEP | |
| CHFR | Q96EP1 | T39 | LLRKREWTIGRRRGC | |
| Chk1 (CHEK1) | O14757 | S280 | AKRPRVTSGGVSESP | - |
| Cip1 (p21, CDKN1A) | P38936 | S145 | GRKRRQTSMTDFYHS | - |
| Cip1 (p21, CDKN1A) | P38936 | S152 | SMTDFYHSKRRLIFS | - |
| Cip1 (p21, CDKN1A) | P38936 | T144 | QGRKRRQTSMTDFYH | - |
| CK1d (CSNK1D) | P48730 | S370 | MERERKVSMRLHRGA | - |
| CLK2 | P49760 | S34 | HKRRRSRSWSSSSDR | + |
| CLK2 | P49760 | T127 | RRRRRSRTFSRSSSQ | + |
| COT (MAP3K8) | P41279 | S400 | EDQPRCQSLDSALLE | + |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| Csdc2 | Q9Y534 | S58 | TKRTRTYSATARASA | |
| EDG-1 | P21453 | T236 | RTRSRRLTFRKNISK | |
| eNOS | P29474 | S1177 | TSRIRTQSFSLQERQ | + |
| EP300 | Q09472 | S1834 | MLRRRMASMQRTGVV | ? |
| ERa (ESR1) | P03372 | S167 | GGRERLASTNDKGSM | ? |
| EZH2 | Q15910 | S21 | CWRKRVKSEYMRLRQ | |
| Ezrin | P15311 | T566 | QGRDKYKTLRQIRQG | |
| FOXO1A | Q12778 | S256 | SPRRRAASMDNNSKF | - |
| FOXO1A | Q12778 | S319 | TFRPRTSSNASTISG | + |
| FOXO1A | Q12778 | T24 | LPRPRSCTWPLPRPE | |
| FOXO3 (FKHRL1) | O43524 | S253 | APRRRAVSMDNSNKY | - |
| FOXO3 (FKHRL1) | O43524 | S315 | DFRSRTNSNASTVSG | - |
| FOXO3 (FKHRL1) | O43524 | T32 | QSRPRSCTWPLQRPE | |
| FOXO4 | P98177 | S197 | APRRRAASMDSSSKL | - |
| FOXO4 | P98177 | S262 | TFRPRSSSNASSVST | - |
| FOXO4 | P98177 | T32 | QSRPRSCTWPLPRPE | - |
| FRAP1 (mTOR) | P42345 | S2448 | RSRTRTDSYSAGQSV | + |
| FRAP1 (mTOR) | P42345 | T2446 | NKRSRTRTDSYSAGQ | - |
| GAB2 | Q9UQC2 | S159 | LLRERKSSAPSHSSQ | - |
| GABRB1 | P18505 | S409 | IQYRKPLSSREAYGR | |
| GABRB2 | P47870 | S434 | SRLRRRASQLKITIP | |
| GATA1 | P15976 | S310 | QTRNRKASGKGKKKR | |
| GATA2 | P23769 | S401 | QTRNRKMSNKSKKSK | |
| Girdin | Q9ULK8 | S1417 | INRERQKSLTLTPTR | |
| Grb10 | Q13322 | S428 | STPVRSVSENSLVAM | |
| Grb10 | Q13322 | S455 | NAPMRSVSENSLVAM | |
| GSK3a | P49840 | S21 | SGRARTSSFAEPGGG | - |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| H3.1 | P68431 | S11 | TKQTARKSTGGKAPR | + |
| HAND2 | P61296 | S114 | KERRRTQSINSAFAE | |
| HMOX-1 | P09601 | S188 | LYRSRMNSLEMTPAV | |
| hnRNP A1 | P09651 | S199 | SQRGRSGSGNFGGGR | - |
| HSP27 | P04792 | S82 | RALSRQLSSGVSEIR | ? |
| HtrA2 | O43464 | S212 | RVRVRLLSGDTYEAV | |
| Huntingtin | P42858 | S421 | GGRSRSGSIVELIAG | |
| IKKa (CHUK) | O15111 | T23 | EMRERLGTGGFGNVC | + |
| IP3R1 (IP3 receptor) | Q14643 | S2690 | FPRMRAMSLVSSDSE | |
| IRAK1 | P51617 | T100 | LRARDIITAWHPPAP | ? |
| IRS1 | P35568 | S270 | EFRPRSKSQSSSNCS | |
| IRS1 | P35568 | S307 | TRRSRTESITATSPA | - |
| IRS1 | P35568 | S330 | SFRVRASSDGEGTMS | |
| IRS1 | P35568 | S527 | RFRKRTHSAGTSPTI | ? |
| KAT6A (MYST3) | Q92794 | T369 | RGRKRKITLSSQSAS | - |
| Mad1 (MXD1) | Q05195 | S145 | IERIRMDSIGSTVSS | - |
| MDM2 | Q00987 | S166 | SSRRRAISETEENSD | |
| MDM2 | Q00987 | S186 | RQRKRHKSDSISLSF | |
| MDM2 | Q00987 | S188 | RKRHKSDSISLSFDE | |
| MDM4 | O15151 | S367 | PDCRRTISAPVVRPK | |
| METTL1 | Q9UBP6 | S27 | YYRQRAHSNPMADHT | - |
| MKK4 (MAP2K4, MEK4) | P45985 | S80 | IERLRTHSIESSGKL | - |
| MLK3 (MAP3K11) | Q16584 | S674 | PGRERGESPTTPPTP | ? |
| MST1 (STK4) | Q13043 | T120 | IIRLRNkTLTEDEIA | - |
| MST1 (STK4) | Q13043 | T387 | TMKRRDETMQPAKPS | + |
| MST2 (CLIK1, STK3) | Q13188 | T117 | IIRLRNkTLIEDEIA | - |
| MST2 (CLIK1, STK3) | Q13188 | T384 | GTMKRNATSPQVQRP | - |
| Myt1 | Q99640 | S75 | ESRPRAVSFRQSEPS | |
| Myt1 | Q99640 | S83 | QLQPRRVSFRGEASE | - |
| NDRG2 | Q9UN36 | T348 | GSRSRSRTLSQSSES | |
| NEDD4L | Q96PU5 | S428 | AEDGASGSATNSNNH | - |
| NuaK1 (ARK5) | O60285 | S600 | PARQRIRSCVSAENF | + |
| Nur77 | P22736 | S351 | GRRGRLPSKPKQPPD | - |
| p27Kip1 | P46527 | S10 | NVRVSNGSPSLERMD | - |
| p27Kip1 | P46527 | T157 | GIRKRPATDDSSTQN | - |
| p27Kip1 | P46527 | T198 | PGLRRRQT_______ | - |
| p47phox | P14598 | S304 | GAPPRRSSIRNAHSI | + |
| p47phox | P14598 | S328 | QDAYRRNSVRFLQQR | + |
| PAK1 | Q13153 | S199 | PRPEHTKSVYTRSVI | + |
| PAK1 | Q13153 | S204 | TKSVYTRSVIEPLPV | + |
| PAK1 | Q13153 | S21 | APPMRNTSTMIGAGS | - |
| Palladin | Q8WX93 | S1118 | VRRPRSRSRDSGDEN | |
| PDCD4 | Q53EL6 | S457 | RGRKRFVSEGDGGRL | |
| PDCD4 | Q53EL6 | S67 | KRRLRKNSSRDSGRG | |
| PDE3A | Q14432 | S292 | VFKRRRRSSSVVSAE | |
| PDE3A | Q14432 | S293 | FKRRRRSSSVVSAEM | |
| PDE3A | Q14432 | S294 | KRRRRSSSVVSAEMS | |
| PDE3B | Q13370 | S295 | VIRPRRRSSCVSLGE | + |
| PEA-15 | Q15121 | S116 | KDIIRQPSEEEIIKL | |
| peripherin | P41219 | S59 | SSSVRLGSFRSPRAG | |
| PFKFB2 | O60825 | S466 | PVRMRRNSFTPLSSS | |
| PFKFB2 | O60825 | S483 | IRRPRNYSVGSRPLK | |
| PIKFYVE | Q9Y2I7 | S318 | LSLDRSGSPMVPSYE | + |
| PIKFYVE (PIP5K) | Q9Y2I7 | S307 | PARNRSASITNLSLD | + |
| PLCG1 | P19174 | S1248 | HGRAREGSFESRYQQ | |
| PRPK (TP53RK) | Q96S44 | S250 | RLRGRKRSMVG____ | + |
| PTP1B (PTPN1) | P18031 | S50 | RNRYRDVSPFDHSRI | + |
| QIK (SIK2) | Q9H0K1 | S358 | DGRQRRPSTIAEQTV | + |
| RAC1 | P63000 | S71 | YDRLRPLSYPQTDVF | - |
| Raf1 | P04049 | S259 | SQRQRSTSTPNVHMV | - |
| RARA | P10276 | S96 | FVCQDKSSGYHYGVS | - |
| Ron (MST1R) | Q04912 | S1394 | VRRPRPLSEPPRPT_ | ? |
| S6 | P62753 | S235 | IAKRRRLSSLRASTS | + |
| S6 | P62753 | S236 | AKRRRLSSLRASTSK | + |
| SLC9A1 (NHE1) | P19634 | S648 | KTRQRLRSYNRHTLV | - |
| SLC9A1 (NHE1) | P19634 | S703 | MSRARIGSDPLAYEP | + |
| SORBS2 | O94875 | S186 | PLRPRDRSSTEKHDW | |
| SORBS2 | O94875 | T188 | RPRDRSSTEKHDWDP | |
| SRPK2 | P78362 | T492 | PSHDRSRTVSASSTG | + |
| SYTL1 | Q8IYJ3 | S237 | PSLDRMLSSSSSVSS | |
| Tal1 | P17542 | T90 | EARHRVPTTELCRPP | |
| Tau | P10636 | S214 | GGKERPGSKEEVDED | |
| TSC2 | P49815 | S1130 | GARDRVRSMSGGHGL | |
| TSC2 | P49815 | S1132 | RDRVRSMSGGHGLRV | |
| TSC2 | P49815 | S939 | SFRARSTSLNERPKS | |
| TSC2 | P49815 | S981 | AFRCRSISVSEHVVR | |
| TSC2 | P49815 | T1462 | GLRPRGYTISDSAPS | |
| VCP | P55072 | S352 | AATNRPNSIDPALRR | |
| VCP | P55072 | S746 | AMRFARRSVSDNDIR | |
| VCP | P55072 | S748 | RFARRSVSDNDIRKY | |
| Wee1 | P30291 | S642 | KKMNRSVSLTIY___ | - |
| Wnk1 (PRKWNK1) | Q9H4A3 | T60 | EYRRRRHTMDKDSRG | + |
| XIAP | P98170 | S87 | VGRHRKVSPNCRFIN | |
| YAP1 | P46937 | S127 | PQHVRAHSSPASLQL | |
| YBX1 | P67809 | S102 | NPRKYLRSVGDGETV | |
| Zyxin | Q15942 | S142 | PQPREKVSSIDLEID |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
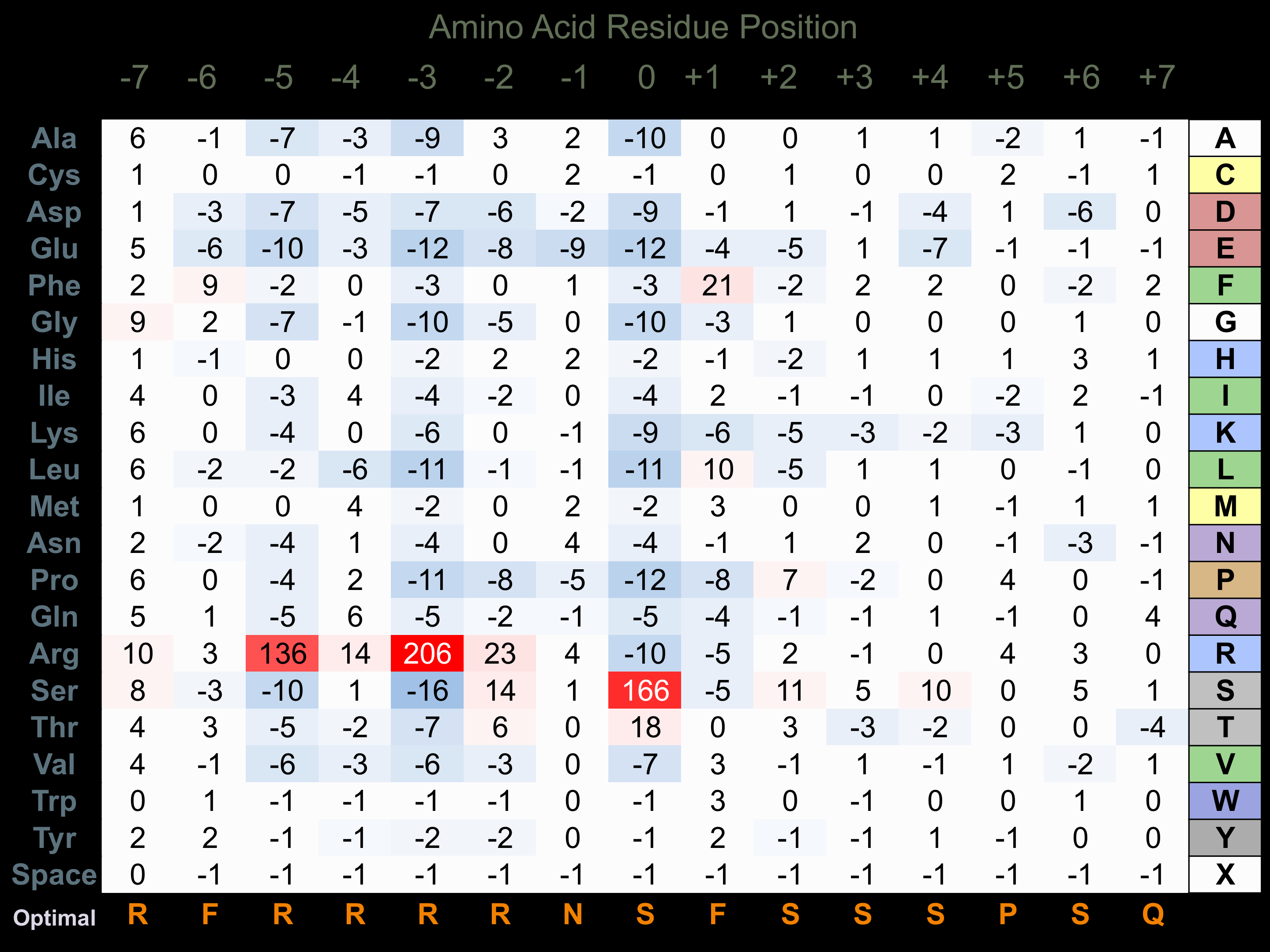
Matrix Type:
Experimentally derived from alignment of 276 known protein substrate phosphosites and 28 peptides phosphorylated by recombinant Akt1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, growth and neurological disorders
Specific Diseases (Non-cancerous):
Proteus Syndrome; Schizophrenia; Leopard Syndrome
Specific Cancer Types:
Breast; Thymoma; Colorectal; Ovarian; Ovarian Serous Cystadenocarcinoma; Cystadenocarcinoma; Anal Squamous Cell Carcinoma; Cowden Syndrome 6; Cowden Syndrome 1; Melanoma; Leukemia
Comments:
AKT1 is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. AKT1 is among the earliest recognized oncoproteins, with a homologue encoded by the v-Akt murine thymoma viral oncogene in the AKT8 retrovirus. up-regulation of PI3K/AKT pathways has been implicated in a variety of cancers. It is primarily mutated in human cancers at the E17, which is located within its PH domain. The E17K mutation is observed in breast cancer, colorectal and ovarian cancer. This somatic mutation results in increased phosphorylation at T308, higher phosphotransferase activity and higher basal ubiquitination. Loss of membrane localization of Akt1 is associated with Q20 mutation. Amplification of AKT1 has been described in human gastric adenocarcinoma, in lung and other cancers.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +69, p<0.001); Brain oligodendrogliomas (%CFC= -51, p<0.012); Breast epithelial cell carcinomas (%CFC= +49, p<0.024); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +458, p<0.0001); Skin fibrosarcomas (%CFC= +105); and Skin melanomas - malignant (%CFC= +954, p<0.0001). The COSMIC website notes an up-regulated expression score for AKT1 in diverse human cancers of 544, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 247 for this protein kinase in human cancers was 4.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.24 % in 46465 diverse cancer specimens. This rate is 3.2-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.62 % in 1428 Meninges cancers tested; 0.81 % in 1945 skin cancers tested; 0.71 % in 353 bone cancers tested; 0.64 % in 4953 breast cancers tested; 0.51 % in 1030 urinary tract cancers tested; 0.46 % in 991 endometrium cancers tested; 0.25 % in 3006 large intestine cancers tested; 0.23 % in 1249 thyroid cancers tested; 0.08 % in 6100 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: E17K (449).
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.

