Nomenclature
Short Name:
ULK1
Full Name:
Serine-threonine-protein kinase ULK1
Alias:
- ATG1
- EC 2.7.1.37
- Unc-51-like kinase 1
- EC 2.7.11.1
- FLJ38455
- UNC51
- Unc51.1
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
ULK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
112,601
# Amino Acids:
1050
# mRNA Isoforms:
1
mRNA Isoforms:
112,631 Da (1050 AA; O75385)
4D Structure:
Interacts with GABARAP and GABARAPL2. Interacts (via C-terminus) with ATG13/KIAA0652. Part of a complex consisting of ATG13/KIAA0652, ULK1 and RB1CC1. Associates with the mammalian target of rapamycin complex 1 (mTORC1) through an interaction with RPTOR;
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
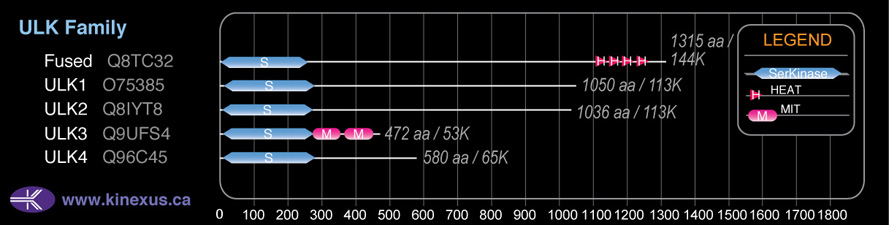
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 16 | 278 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S87, S111, S131, S158, S195, S224, S225, S341, S403, S405, S409, S415, S429, S450, S459, S460, S465, S467, S469, S477, S479, S495, S533, S538, S539, S544, S556+, S561, S583, S588, S593, S598, S605, S623, S638, S639, S694, S716, S719, S748, S758, S761, S775, S781, S866, S912, S949, S1042.
Threonine phosphorylated:
T180+, T456, T468, T575, T625, T636, T654, T660, T695, T699, T709, T717, T764, T1046.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 22
22
740
19
883
 5
5
163
10
117
 2
2
81
1
0
 13
13
417
46
563
 20
20
670
14
657
 1.3
1.3
42
37
26
 11
11
375
19
683
 22
22
714
17
658
 8
8
279
10
240
 5
5
171
45
196
 4
4
142
14
109
 24
24
779
95
649
 1.2
1.2
39
12
10
 5
5
158
9
114
 6
6
213
11
205
 5
5
162
8
141
 6
6
192
102
106
 3
3
87
8
48
 5
5
177
60
220
 18
18
599
56
632
 4
4
144
10
127
 7
7
224
12
250
 0.2
0.2
6
2
2
 3
3
112
8
107
 7
7
234
10
280
 21
21
694
24
657
 1
1
32
15
16
 3
3
93
8
65
 6
6
184
8
151
 4
4
123
14
98
 34
34
1123
18
790
 100
100
3291
27
4630
 3
3
85
53
197
 23
23
767
26
650
 7
7
240
22
627
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 84.2
84.2
86.1
92 89.1
89.1
89.7
98 -
-
-
90 -
-
-
- 87.9
87.9
91.2
89 -
-
-
- 89.8
89.8
92.7
90 -
-
-
90 -
-
-
- 52.1
52.1
63.9
- 22.4
22.4
31.8
77 22
22
31.7
71 57.3
57.3
69.1
64.5 -
-
-
- 34.3
34.3
47.7
- 36.6
36.6
49.3
- 33.6
33.6
47.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 20
20
35.1
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | GABARAP - O95166 |
| 2 | GABARAPL2 - P60520 |
| 3 | SYNGAP1 - Q96PV0 |
| 4 | OCLN - Q16625 |
| 5 | SDCBP - O00560 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AMPKa1 (PRKAA1) | Q13131 | T368 | FLDDHHLTRPHPERV | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
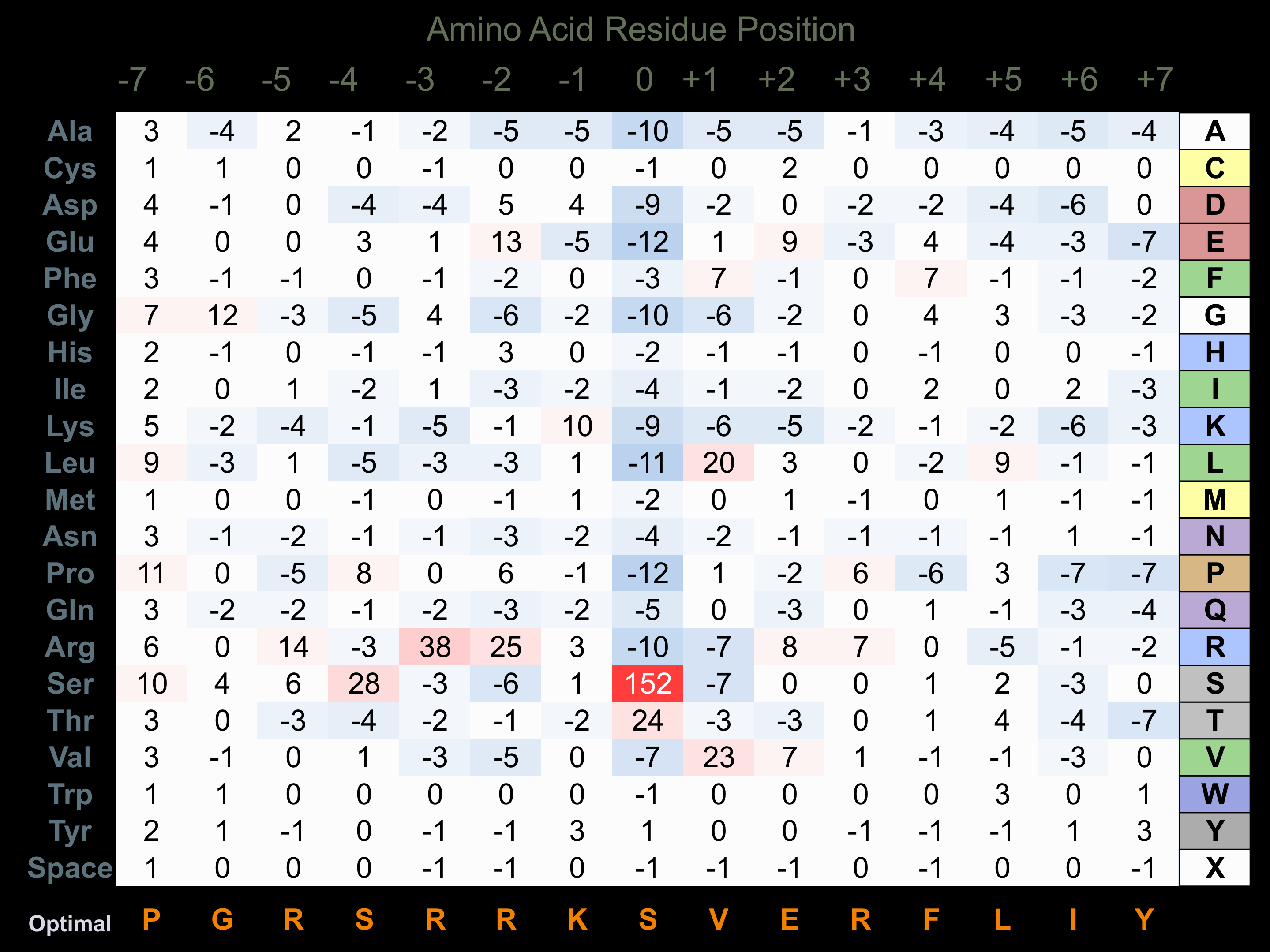
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Gastrointestinal disorder
Specific Diseases (Non-cancerous):
Crohn's disease
Comments:
The pathogenesis of Crohn's disease has been associated with an aberrant autophagic response through an association with the ATG16L1 and IRGM genes, which are involved in autophagic regulation. ULK1 is a downstream effector and negative regulator of both mTOR (via interaction with RPTOR) and AMPK, thus the protein is a key component of the regulatory feedback loops present in autophagy. Crohn's disease is a gastrointestinal disease characterized by excessive and inappropriate inflammation in the gastrointestinal tract. Primary symptoms of the disease include diarrhea, abdominal pain, fever, and weight loss, which can be accompanied by anemia, skin irritation, eye inflammation, and general tiredness. Due to its role in regulating autophagy, ULK1 has been hypothesized to be a suceptibility gene for Crohn's disease. A significantly higher frequency of a particular single nucleotide polymorphism (SNP), the rs12303764(T) allele, was observed in patients with Crohn's disease as compared to controls, indicating that genetic variation in ULK1 may contribute to Crohn's disease suceptibility.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Colorectal adenocarcinomas (early onset) (%CFC= +81, p<0.004); Oral squamous cell carcinomas (OSCC) (%CFC= +79, p<0.015); Pituitary adenomas (ACTH-secreting) (%CFC= +337); Prostate cancer (%CFC= +49, p<0.015); and Skin squamous cell carcinomas (%CFC= +67, p<0.053). The COSMIC website notes an up-regulated expression score for ULK1 in diverse human cancers of 471, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 34 for this protein kinase in human cancers was 0.6-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24726 diverse cancer specimens. This rate is only -1 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.37 % in 864 skin cancers tested; 0.33 % in 1270 large intestine cancers tested; 0.26 % in 589 stomach cancers tested; 0.17 % in 603 endometrium cancers tested; 0.15 % in 1634 lung cancers tested; 0.14 % in 548 urinary tract cancers tested; 0.11 % in 710 oesophagus cancers tested; 0.07 % in 1512 liver cancers tested; 0.07 % in 127 biliary tract cancers tested; 0.05 % in 833 ovary cancers tested; 0.04 % in 1316 breast cancers tested; 0.04 % in 1276 kidney cancers tested; 0.03 % in 558 thyroid cancers tested; 0.03 % in 273 cervix cancers tested; 0.03 % in 1459 pancreas cancers tested; 0.02 % in 881 prostate cancers tested; 0.02 % in 2082 central nervous system cancers tested; 0.01 % in 942 upper aerodigestive tract cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: A125T (3).
Comments:
Thirteen deletions, no insertions and 1 complex mutation are noted on the COSMIC website.

