Nomenclature
Short Name:
AMPKa1
Full Name:
5'-AMP-activated protein kinase, catalytic alpha-1 chain
Alias:
- 5'-AMP-activated protein kinase, catalytic alpha-1 chain
- AMPK-alpha1
- HMG-CoA reductase kinase
- HMG-CoA reductase kinase
- PRKAA1
- Protein kinase, AMP-activated, alpha 1 catalytic subunit
- AAPK1
- Acetyl-CoA carboxylase kinase
- AMPK alpha-1 chain
- AMPK, alpha, 1
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMKL
SubFamily:
AMPK
Specific Links
Structure
Mol. Mass (Da):
64009
# Amino Acids:
559
# mRNA Isoforms:
2
mRNA Isoforms:
65,523 Da (574 AA; Q13131-2); 64,009 Da (559 AA; Q13131)
4D Structure:
Heterotrimer of an alpha catalytic subunit, a beta and a gamma non-catalytic subunits. Interacts with FNIP1 and FNIP2.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 27 | 279 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K80.
Serine phosphorylated:
S6+, S172, S176, S184+, S187-, S293, S356, S360, S397, S467, S486, S487, S494, S496, S498, S506, S508, S520, S523, S524, S527, S531.
Threonine phosphorylated:
T32, T183+, T269+, T355, T368-, T382, T388, T482, T488, T490, T522, T526.
Tyrosine phosphorylated:
Y190, Y247, Y294, Y441, Y442, Y463, Y500.
Ubiquitinated:
K285, K396, K485.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 33
33
1378
47
1176
 0.5
0.5
20
21
17
 4
4
176
10
130
 6
6
232
163
707
 17
17
696
55
539
 0.8
0.8
33
105
55
 3
3
117
64
259
 16
16
672
45
1412
 9
9
364
20
348
 2
2
90
135
93
 3
3
117
34
138
 13
13
538
196
521
 2
2
95
32
166
 0.5
0.5
20
15
13
 1
1
41
21
47
 0.7
0.7
31
30
28
 3
3
106
254
116
 2
2
92
20
118
 1.2
1.2
52
124
48
 14
14
589
191
521
 2
2
94
27
107
 2
2
105
31
113
 3
3
136
20
169
 6
6
262
22
218
 2
2
95
28
106
 20
20
857
106
913
 2
2
65
38
108
 3
3
126
22
147
 3
3
109
21
139
 3
3
123
56
186
 20
20
823
24
740
 100
100
4218
51
10863
 4
4
179
139
580
 19
19
815
135
724
 14
14
598
74
1289
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 97.2
97.2
97.4
99.5 -
-
-
99 -
-
-
98 92.5
92.5
93.8
99 -
-
-
- 98.8
98.8
99.5
99 99.1
99.1
99.6
99 -
-
-
- 89.2
89.2
89.2
- 27.1
27.1
43
95.5 88.7
88.7
94.6
90 84.4
84.4
90.9
88 -
-
-
- -
-
-
- 53.5
53.5
62.2
- -
-
53.5
58 -
-
-
- -
-
-
- -
-
-
- -
-
-
52 45.1
45.1
62.4
57 33.5
33.5
52
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PRKAB1 - Q9Y478 |
| 2 | TSC2 - P49815 |
| 3 | PRKAG1 - P54619 |
| 4 | PRKAB2 - O43741 |
| 5 | PRKAG3 - Q9UGI9 |
| 6 | CFTR - P13569 |
| 7 | CAB39 - Q9Y376 |
| 8 | RAF1 - P04049 |
| 9 | EEF2K - O00418 |
| 10 | RAPGEF6 - Q8TEU7 |
| 11 | NUAK1 - O60285 |
| 12 | CDC37 - Q16543 |
| 13 | HSPA5 - P11021 |
| 14 | PRKAG2 - Q9UGJ0 |
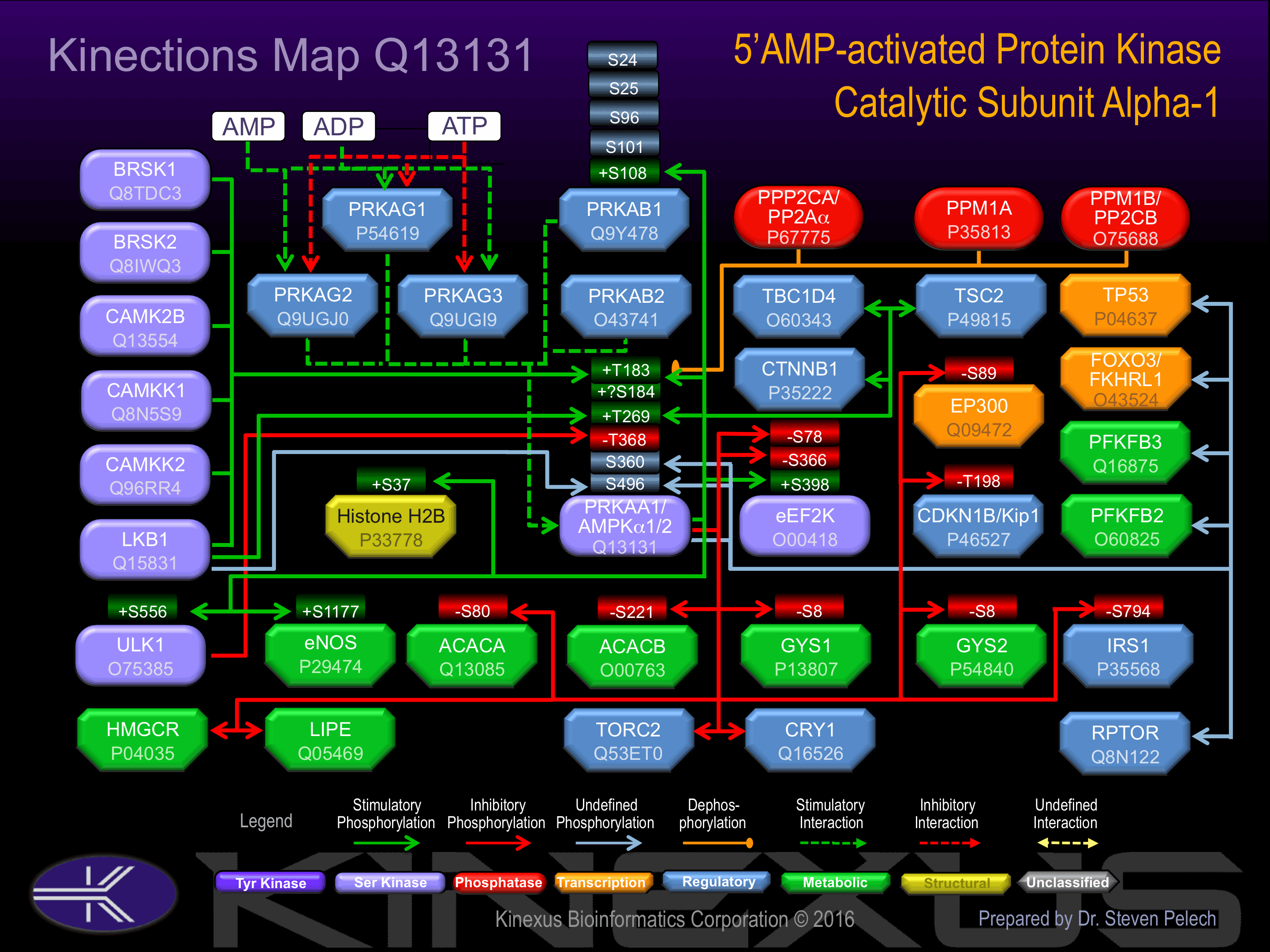
Regulation
Activation:
Binding of AMP results in allosteric activation, inducing phosphorylation on Thr-183 by STK11 in complex with STE20-related adapter-alpha (STRAD alpha) pseudo kinase and CAB39. Phosphorylation at Thr-174 by CAMKK1 and CAMKK2 increases phosphotransferase activity, triggered by a rise in intracellular calcium ions, without detectable changes in the AMP/ATP ratio.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| BRSK2 | Q8IWQ3 | T183 | SDGEFLRTSCGSPNY | + |
| BRSK1 | Q8TDC3 | T183 | SDGEFLRTSCGSPNY | + |
| AMPKa1 | Q13131 | T183 | SDGEFLRTSCGSPNY | + |
| LKB1 | Q15831 | T183 | SDGEFLRTSCGSPNY | + |
| CaMK2b | Q13554 | T183 | SDGEFLRTSCGSPNY | + |
| CaMKK2 | Q96RR4 | T183 | SDGEFLRTSCGSPNY | + |
| CaMKK1 | Q8N5S9 | T183 | SDGEFLRTSCGSPNY | + |
| AMPKa1 | Q13131 | T269 | VDPMKRATIKDIREH | + |
| LKB1 | Q15831 | T269 | VDPMKRATIKDIREH | + |
| ULK1 | O75385 | T368 | FLDDHHLTRPHPERV | - |
| PKACa | P17612 | S487 | ATPQRSGSVSNYRSC | |
| LKB1 | Q15831 | S496 | ATPQRSGSVSNYRSC |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ACACA | Q13085 | S80 | LHIRSSMSGLHLVKQ | - |
| ACACB | O00763 | S221 | PTMRPSMSGLHLVKR | - |
| AMPKa1 (PRKAA1) | Q13131 | S360 | LATSPPDSFLDDHHL | |
| AMPKa1 (PRKAA1) | Q13131 | S496 | ATPQRSGSVSNYRSC | |
| AMPKa1 (PRKAA1) | Q13131 | T183 | SDGEFLRTSCGSPNY | + |
| AMPKa1 (PRKAA1) | Q13131 | T269 | VDPMKRATIKDIREH | + |
| AMPKb1 | Q9Y478 | S101 | SGSFNNWSKLPLTRS | |
| AMPKb1 | Q9Y478 | S108 | SKLPLTRSHNNFVAI | + |
| AMPKb1 | Q9Y478 | S24 | HKTPRRDSSGGTKDG | |
| AMPKb1 | Q9Y478 | S25 | KTPRRDSSGGTKDGD | |
| AMPKb1 | Q9Y478 | S96 | KEVYLSGSFNNWSKL | |
| ChREBP | Q9NP71 | S556 | TLLRSPGSPQETVPE | |
| CK1e1 (CSNK1E) | P49674 | S389 | RGAPANVSSSDLTGR | - |
| eEF2K | O00418 | S366 | SPRVRTLSGSRPPLL | - |
| eEF2K | O00418 | S398 | DSLPSSPSSATPHSQ | + |
| eEF2K | O00418 | S78 | SSGSPANSFHFKEAW | - |
| eNOS | P29474 | S1177 | TSRIRTQSFSLQERQ | + |
| eNOS | P29474 | T495 | TGITRKKTFKEVANA | |
| EP300 | Q09472 | S89 | SELLRSGSSPNLNMG | - |
| FRAP1 (mTOR) | P42345 | T2446 | NKRSRTRTDSYSAGQ | - |
| GABBR2 | O75899 | S784 | VTSVNQASTSRLEGL | |
| GYS1 | P13807 | S8 | MPLNRTLSMSSLPGL | - |
| GYS2 | P54840 | S8 | MLRGRSLSVTSLGGL | - |
| Histone H2B | P33778 | S37 | RKRSRKESYSIYVYK | + |
| HMG-CoA reductase | P04035 | S872 | SHMIHNRSKINLQDL | |
| HNF4A | P41235 | S313 | GKIKRLRSQVQVSLE | |
| HSL | Q05469 | S554 | EPMRRSVSEAALAQP | |
| IRS1 | P35568 | S794 | QHLRLSTSSGRLLYA | - |
| KPNA2 | P52292 | S105 | QAARKLLSREKQPPI | |
| NM23 (NME1) | P15531 | S122 | NIIHGSDSVESAEKE | |
| NM23 (NME1) | P15531 | S144 | EELVDYTSCAQNWIY | |
| nNOS | P29475 | S1417 | TNRLRSESIAFIEES | |
| p27Kip1 | P46527 | T198 | PGLRRRQT_______ | - |
| PFKFB2 | O60825 | S466 | PVRMRRNSFTPLSSS | |
| PFKFB3 | Q16875 | S461 | NPLMRRNSVTPLASP | |
| Raptor | Q8N122 | S792 | DKMRRASSYSSLNSL | |
| TSC2 | P49815 | S1387 | QPLSKSSSSPELQTL | |
| TSC2 | P49815 | T1271 | PPLPRSNTVASFSSL | |
| ULK1 | O75385 | S556 | GLGCRLHSAPNLSDL | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
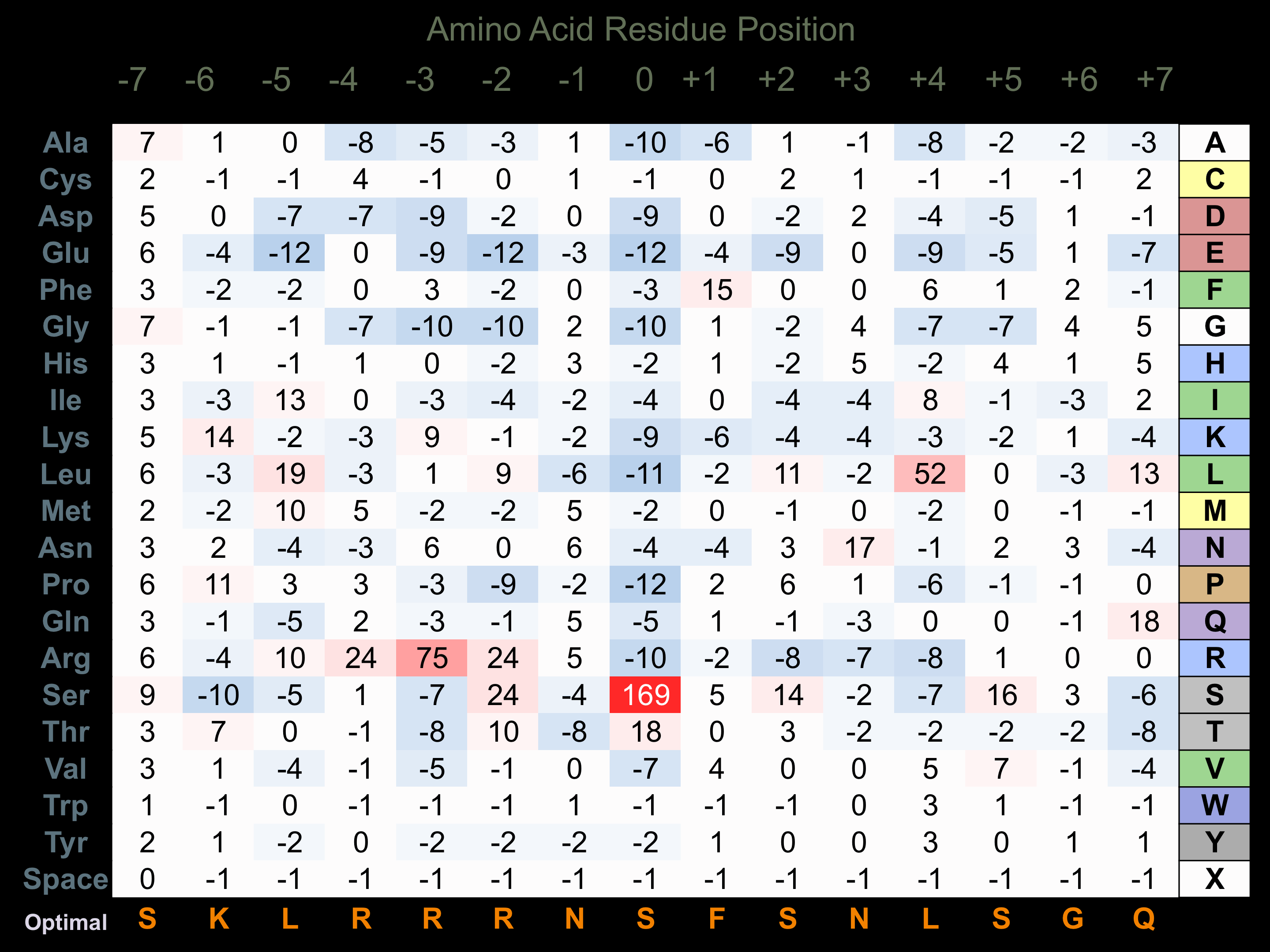
Matrix Type:
Experimentally derived from alignment of 104 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cardiovascular disorders
Specific Diseases (Non-cancerous):
Wolff-Parkinson-White (WPW) syndrome
Comments:
Wolff-Parkinson-White syndrome (WPW), also known as anomalous atrioventricular excitation, is a cardiovascular disease characterized by an abnormal heartbeat (arrhythmia) resulting from the uncoordinated conduction of electrical signals through the myocardium. This disease is one of the most common causes of rapid heart rate (tachycardia) in infants and children. Although the cause in most cases is unknown, autosomal dominant mutations in AMPKa1 have been implicated in the disorder. AMPKa1 is the catalytic subunit of AMP-activated protein kinase (AMPK), a protein kinase that functions as a cellular energy sensor and plays a key role in the regulation of cellular metabolism. When cellular ATP levels drop (e.g. during starvation situations), AMPK simultaneously promotes energy producing mechanisms and inhibits energy-consuming pathways, such as metabolite biosynthesis and cellular proliferation/growth. In general, several mutations in AMPK have been associated with abnormal cardiac function in humans, specifically ventricular hypertrophy and electrophysiological abnormalities, which are also characteristic of WPW syndrome. None of the observed mutations were correlated with changes in the phosphorylation state of the T172 residue of AMPK, but resulted in either a reduced AMP-dependence of protein activity or reduced AMP sensitivity. Transgenic mice overexpressing a mutant AMPK?2 subunit (N488I substitution mutation) displayed increased catalytic activity of AMPK, a 30-fold increase in accumulation of cardiac glycogen, left ventricular hypertrophy, and ventricular pre-excitation and dysfunction of the sino-atiral node. In addition, electrophysiological recordings revealed alternative pathways for electrical conduction through the atrial and ventricular musculature, consistent with the phenotype of WPW syndrome. Histological analysis of the tissue demonstrated that glycogen-filled cardiomyocytes disrupted the annulus fibrosis, which is an insulating layer that prevents the inappropriate electrical stimulation of the ventricles by atrial electrical activity. The loss of tissue integrity in the annulus fibrosis is thought to be the anatomical substrate mediating the pre-excitation of the ventricles, thus potentially explaining the symptoms of WPW syndrome. Therefore, aberrant activity of AMPK may play a important role in the development of WPW syndrome.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -61, p<0.052); Brain oligodendrogliomas (%CFC= -74, p<0.023); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +525, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= +45, p<0.033); and Prostate cancer - primary (%CFC= +50, p<0.0002). The COSMIC website notes an up-regulated expression score for AMPKa1 in diverse human cancers of 712, which is 1.5-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 95 for this protein kinase in human cancers was 1.6-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.04 % in 25389 diverse cancer specimens. This rate is -44 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.19 % in 1119 large intestine cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,655 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.