Nomenclature
Short Name:
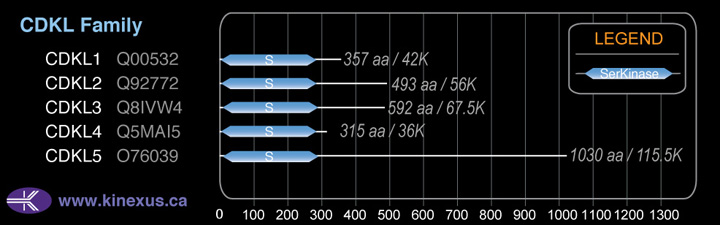
CDKL5
Full Name:
Serine-threonine-protein kinase 9
Alias:
- Cyclin-dependent kinase-like 5
- EC 2.7.11.22
- Kinase CdkL5
- STK9
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDKL
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
115,538
# Amino Acids:
1030
# mRNA Isoforms:
2
mRNA Isoforms:
115,538 Da (1030 AA; O76039); 107,519 Da (960 AA; O76039-2)
4D Structure:
Interacts with MECP2.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 13 | 297 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K2, K12, K33.
Serine phosphorylated:
S306, S308, S341, S343, S377, S407, S446, S479, S488, S493, S512, S520, S526, S529, S543, S603, S646, S681, S720, S726, S761, S833, S987.
Threonine phosphorylated:
T169+, T533, T658, T813, T831, T983.
Tyrosine phosphorylated:
Y168, Y171+, Y177, Y188, Y262, Y286, Y482, Y686, Y704.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 56
56
458
29
903
 3
3
21
17
17
 48
48
390
20
375
 59
59
482
102
785
 33
33
264
23
203
 5
5
43
79
158
 41
41
334
35
607
 76
76
615
53
813
 26
26
213
17
165
 15
15
118
104
174
 14
14
112
43
138
 60
60
487
192
471
 17
17
141
42
202
 3
3
24
14
20
 31
31
251
37
372
 1.2
1.2
10
14
12
 11
11
90
215
193
 30
30
244
31
276
 8
8
65
101
93
 29
29
237
109
278
 38
38
306
34
404
 16
16
132
39
170
 32
32
263
30
309
 94
94
764
31
1245
 21
21
167
35
233
 90
90
733
67
1163
 18
18
148
45
237
 35
35
287
30
702
 40
40
325
31
475
 13
13
109
28
74
 32
32
258
24
258
 100
100
811
36
1419
 1
1
8
67
58
 75
75
608
57
577
 30
30
243
35
307
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
99 26.7
26.7
39.5
97.5 -
-
-
98 -
-
-
97 88.1
88.1
89.7
98 -
-
-
- 85.9
85.9
88.9
96 23.6
23.6
36.5
96 -
-
-
- 76.5
76.5
82.5
- 75.2
75.2
83.6
83.5 -
-
-
76 55.3
55.3
69.1
68 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 25.5
25.5
39
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Likely that phosphorylation at Tyr-171 is required for catalytic activity based on homology with other CDK's.
Inhibition:
Likely that phosphorylation at Tyr-24 is inhibitory based on homology with other CDK's.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
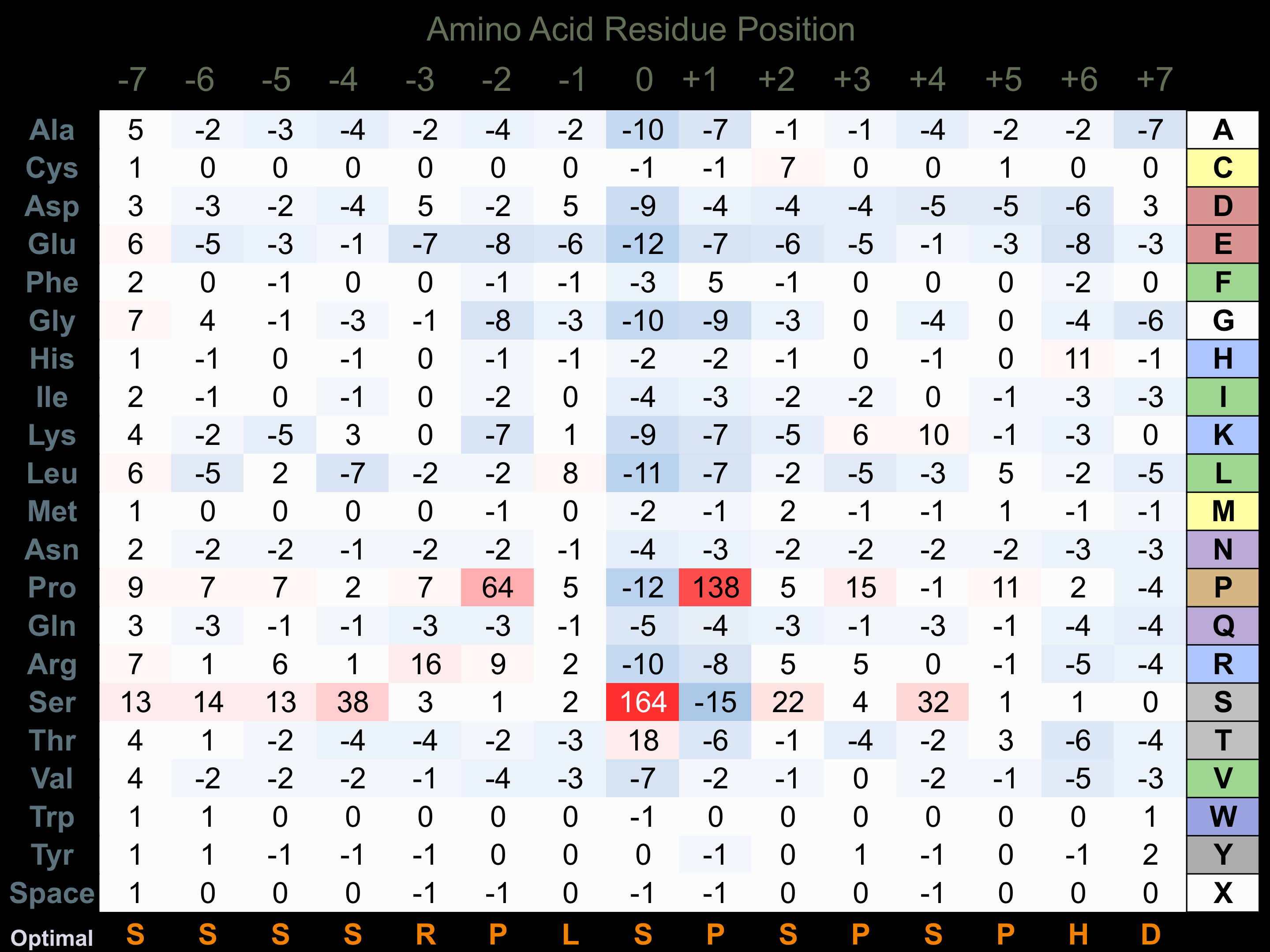
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AC1NS7CD | Kd = 1.7 nM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 1.7 nM | 3025986 | 296468 | 22037378 |
| AT7519 | Kd = 2.6 nM | 11338033 | 22037378 | |
| AST-487 | Kd = 6.6 nM | 11409972 | 574738 | 22037378 |
| Alvocidib | Kd = 7.1 nM | 9910986 | 428690 | 22037378 |
| Lestaurtinib | Kd = 10 nM | 126565 | 22037378 | |
| A674563 | Kd = 200 nM | 11314340 | 379218 | 22037378 |
| Staurosporine | Kd = 340 nM | 5279 | 22037378 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Foretinib | Kd = 2.7 µM | 42642645 | 1230609 | 22037378 |
| Tozasertib | Kd = 2.8 µM | 5494449 | 572878 | 22037378 |
| R547 | Kd = 4 µM | 6918852 | 22037378 | |
| Ruboxistaurin | Kd = 4.1 µM | 153999 | 91829 | 22037378 |
Disease Linkage
General Disease Association:
Cancer, neurological, and developmental disorders
Specific Diseases (Non-cancerous):
Rett syndrome; West syndrome; Epileptic encephalopathy Early infantile Type 2 (EIEE2); Angelman syndrome-Like; Seizure disorder; Angelman syndrome; Aicardi syndrome; Gait apraxia; Epileptic encephalopathy, Early infantile, 5; Epileptic encephalopathy, Early infantile, 17; CDKL5-related Angelman-like syndrome
Comments:
EIEE2 sufferers experience seizures, spasms, and impaired motor control early in life. In EIEE2 protein mislocation (to the cytoplasm) can occur with an A40V, or L220P mutation. Mislocation can occur and protein activity can be affected with the C152F, and R175S mutations. Rett Syndrome is a neuro-development disorder affecting psychomotor development, autistic like features, seizures, and slow head growth. Rett Syndrome is also associated with neuroblastomas. West seizure is a form of epilepsy in infancy. Cdkl5-Related Disorder results in epilepsy and severe intellectual dissability. Atypical Rett Syndrome sufferers will have either a milder or more extreme version of Rett Syndrome. Aicardi Syndrome can include cleft lip and retinal detachment.
Specific Cancer Types:
Rett syndrome
Comments:
CDKL5 has been linked with Rett Syndrome, which is a neuro-development disorder affecting psychomotor development, autistic like features, seizures, and slow head growth. Rett Syndrome is also associated with neuroblastomas.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Brain glioblastomas (%CFC= -69, p<0.018). The COSMIC website notes an up-regulated expression score for CDKL5 in diverse human cancers of 355, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 14 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24790 diverse cancer specimens. This rate is very similar (+ 5% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.37 % in 1296 large intestine cancers tested; 0.31 % in 589 stomach cancers tested; 0.27 % in 603 endometrium cancers tested; 0.25 % in 864 skin cancers tested; 0.21 % in 273 cervix cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S603F (8); R875W (5).
Comments:
Only 2 insertions, and no deletions or complex mutations are noted on the COSMIC website.

