Nomenclature
Short Name:
PKD3
Full Name:
Protein kinase C, nu type
Alias:
- EC 2.7.11.13
- PRKD3
- Protein kinase C, nu type
- Protein kinase D3
- Protein kinase EPK2
- EPK2
- KPCD3
- NPKC-nu
- PRKCN
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
PKD
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
100,471
# Amino Acids:
890
# mRNA Isoforms:
2
mRNA Isoforms:
100,471 Da (890 AA; O94806); 68,123 Da (611 AA; O94806-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
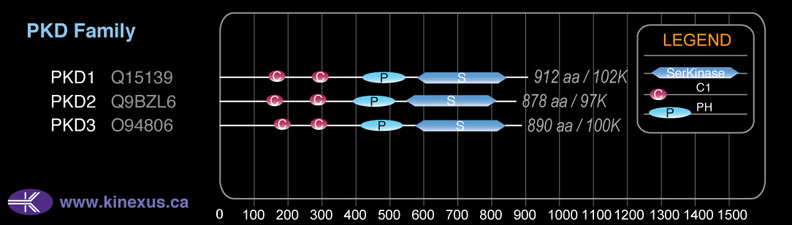
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S2, S6, S9, S27, S30, S31, S37, S41, S44, S46, S213, S216, S252, S254, S364, S391, S395, S415, S543, S731+, S735+.
Threonine phosphorylated:
T34, T360, T389, T535+, T739-.
Tyrosine phosphorylated:
Y742-, Y852, Y867.
Ubiquitinated:
K293, K730, K816.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 81
81
1193
47
914
 5
5
71
20
38
 7
7
98
19
47
 30
30
442
185
1396
 52
52
766
48
557
 5
5
80
107
115
 12
12
172
65
366
 100
100
1474
65
3166
 31
31
460
17
359
 5
5
69
152
65
 3
3
48
43
35
 43
43
634
215
632
 10
10
143
41
69
 5
5
76
11
71
 3
3
41
20
43
 4
4
62
31
77
 3
3
50
286
45
 5
5
78
28
51
 5
5
69
136
64
 50
50
739
188
613
 5
5
77
37
57
 11
11
159
39
101
 8
8
117
21
54
 12
12
178
30
126
 7
7
97
38
81
 44
44
648
124
738
 10
10
147
44
80
 5
5
72
28
39
 10
10
141
28
88
 2
2
24
56
21
 40
40
584
24
439
 35
35
515
51
648
 59
59
873
105
1473
 51
51
758
130
621
 21
21
312
74
622
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 97.3
97.3
97.3
100 99.5
99.5
99.6
100 -
-
-
98 -
-
-
99 97.1
97.1
98.7
97 -
-
-
- 95.7
95.7
97.8
96 68.4
68.4
79
96 -
-
-
- 76.4
76.4
79.7
- -
-
-
90 -
-
-
- 82
82
89.4
84 -
-
-
- -
-
-
59 -
-
-
- 45.1
45.1
57.8
56 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | KPNB1 - Q14974 |
| 2 | VAMP2 - P63027 |
| 3 | KPNA2 - P52292 |
| 4 | GSK3A - P49840 |
Regulation
Activation:
Activated by diacylglycerol (DAG) and phorbol esters. Phorbol-ester/DAG-type domains 1 and 2 bind both DAG and phorbol ester with high affinity and mediate translocation to the cell membrane. Activated by phosphorylation at Ser-731 and Ser-735.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
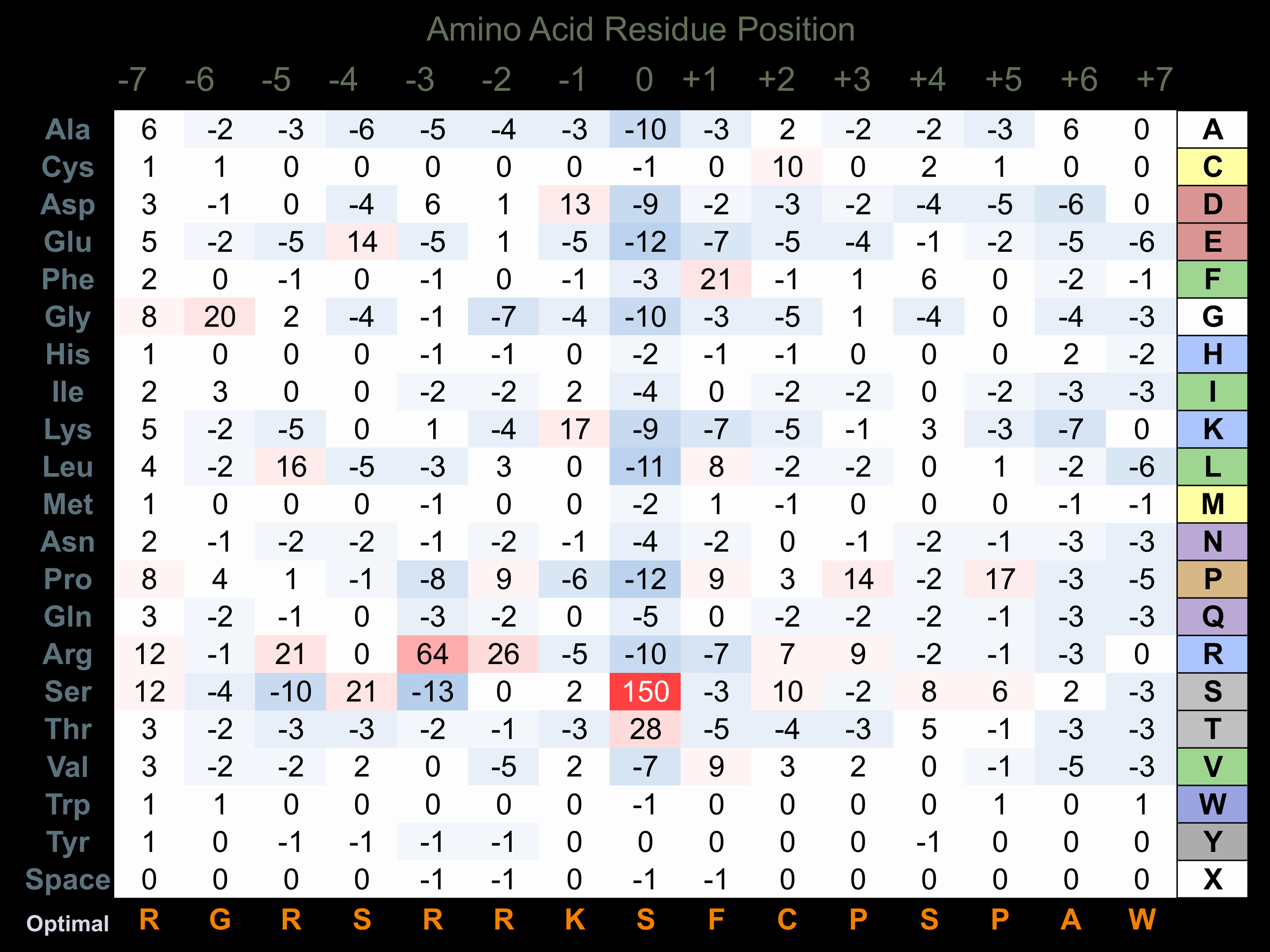
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Chronic lymphocytic leukemia; Melanomas; Lung cancer; Prostate Cancer
Comments:
Mutations of the PKD3 gene were detected in some cases of chronic lymphocytic leukemia. It can also sensitize the effects of Raf kinase inhibitors in melanoma cells. It may be involve in the metastasis of lung cancer and contribute to prostate cancer cell growth.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +75, p<0.0001); Brain glioblastomas (%CFC= -62, p<0.015); Brain oligodendrogliomas (%CFC= -55, p<0.03); Breast epithelial carcinomas (%CFC= -51, p<0.062); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -46, p<0.06); Clear cell renal cell carcinomas (cRCC) (%CFC= +59, p<0.053); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -76, p<0.0001); Pituitary adenomas (aldosterone-secreting) (%CFC= +84, p<0.027); Prostate cancer - primary (%CFC= +48, p<0.0002); and Skin melanomas - malignant (%CFC= +173, p<0.0001). The COSMIC website notes an up-regulated expression score for PKD3 in diverse human cancers of 432, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 53 for this protein kinase in human cancers was 0.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24936 diverse cancer specimens. This rate is only -13 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.12 % in 10 peritoneum cancers tested; 0.29 % in 1270 large intestine cancers tested; 0.18 % in 864 skin cancers tested; 0.17 % in 589 stomach cancers tested; 0.16 % in 942 upper aerodigestive tract cancers tested; 0.15 % in 603 endometrium cancers tested; 0.14 % in 548 urinary tract cancers tested; 0.1 % in 1316 breast cancers tested; 0.08 % in 273 cervix cancers tested; 0.08 % in 1634 lung cancers tested; 0.07 % in 1512 liver cancers tested; 0.05 % in 1276 kidney cancers tested; 0.04 % in 833 ovary cancers tested; 0.04 % in 558 thyroid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: N796K (6).
Comments:
Only 4 deletions, no insertions or complex mutations are noted on the COSMIC website.

