Nomenclature
Short Name:
ARAF
Full Name:
A-Raf proto-oncogene serine-threonine-protein kinase
Alias:
- ARAF
- Kinase A-Raf
- PKS
- PKS2
- Proto-oncogene Pks
- V-raf murine sarcoma 3611 viral oncogene
- A-RAF
- ARAF1
- A-raf-1
- EC 2.7.11.1
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
RAF
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
67,585
# Amino Acids:
606
# mRNA Isoforms:
2
mRNA Isoforms:
67,585 Da (606 AA; P10398); 20,904 Da (186 AA; P10398-2)
4D Structure:
Interacts with TH1L/NELFD
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
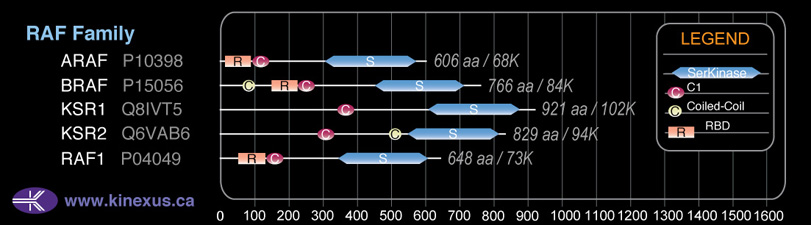
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Other:
R524 (Omega-N-methylarginine (predicted).
Serine phosphorylated:
S15, S157, S162, S186, S212, S214+, S222, S229, S250, S257+, S259, S262+, S264+, S265, S269, S272, S274, S283, S299, S320-, S341, S432+, S458+, S528, S532, S535, S536, S547, S573, S580, S582+, S585, S600.
Threonine phosphorylated:
T20, T181, T213, T215, T223, T224, T253, T318, T323, T344, T413, T442+, T452+, T455+, T589.
Tyrosine phosphorylated:
Y24, Y42, Y155, Y296, Y301+, Y302+, Y419, Y526.
Ubiquitinated:
K314, K454, K540.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 39
39
1382
22
1281
 7
7
246
13
157
 10
10
337
16
265
 20
20
699
94
1538
 20
20
706
24
673
 38
38
1323
61
3188
 15
15
509
33
757
 16
16
564
46
558
 19
19
679
10
592
 12
12
410
100
456
 13
13
448
35
337
 20
20
690
150
589
 10
10
351
27
238
 6
6
217
12
156
 11
11
402
32
301
 4
4
145
14
135
 8
8
281
287
223
 10
10
349
25
274
 13
13
467
89
420
 18
18
634
84
568
 7
7
237
31
156
 13
13
462
33
353
 13
13
445
33
284
 6
6
223
25
256
 10
10
341
31
271
 18
18
619
65
657
 9
9
331
30
299
 13
13
443
25
498
 9
9
303
25
231
 11
11
396
28
391
 31
31
1087
18
622
 100
100
3508
26
6647
 3
3
105
66
320
 21
21
740
62
631
 4
4
146
35
199
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 58.2
58.2
70.8
0 97
97
97.7
96 -
-
-
97 -
-
-
97 92.7
92.7
94.1
97 -
-
-
- 95.2
95.2
97
95 95.5
95.5
97
96 -
-
-
- 47.7
47.7
57.7
- 58.1
58.1
71.3
- 57.8
57.8
70.7
- 67.4
67.4
77.8
68 63.5
63.5
74.3
- 34.5
34.5
48.9
- -
-
-
- 32.8
32.8
48.1
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TH1L - Q8IXH7 |
| 2 | MAP2K1 - Q02750 |
| 3 | PIK3R1 - P27986 |
| 4 | RRAS - P10301 |
| 5 | PRPF6 - O94906 |
| 6 | TIMM44 - O43615 |
| 7 | LY6G5B - Q8NDX9 |
| 8 | CSNK2B - P67870 |
| 9 | HRAS - P01112 |
| 10 | YWHAH - Q04917 |
| 11 | MAP2K2 - P36507 |
| 12 | PDGFRB - P09619 |
| 13 | CSNK2B - P67870 |
| 14 | PBK - Q96KB5 |
| 15 | ASS1 - P00966 |
Regulation
Activation:
Phosphorylation of Ser-214, Ser-257, Ser-262, Ser-264, Tyr-301, Tyr-302, Ser-432, Thr-442, Thr-452, Thr-455 and Ser-582 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| MEK1 (MAP2K1) | Q02750 | S218 | VSGQLIDSMANSFVG | + |
| MEK1 (MAP2K1) | Q02750 | S222 | LIDSMANSFVGTRSY | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
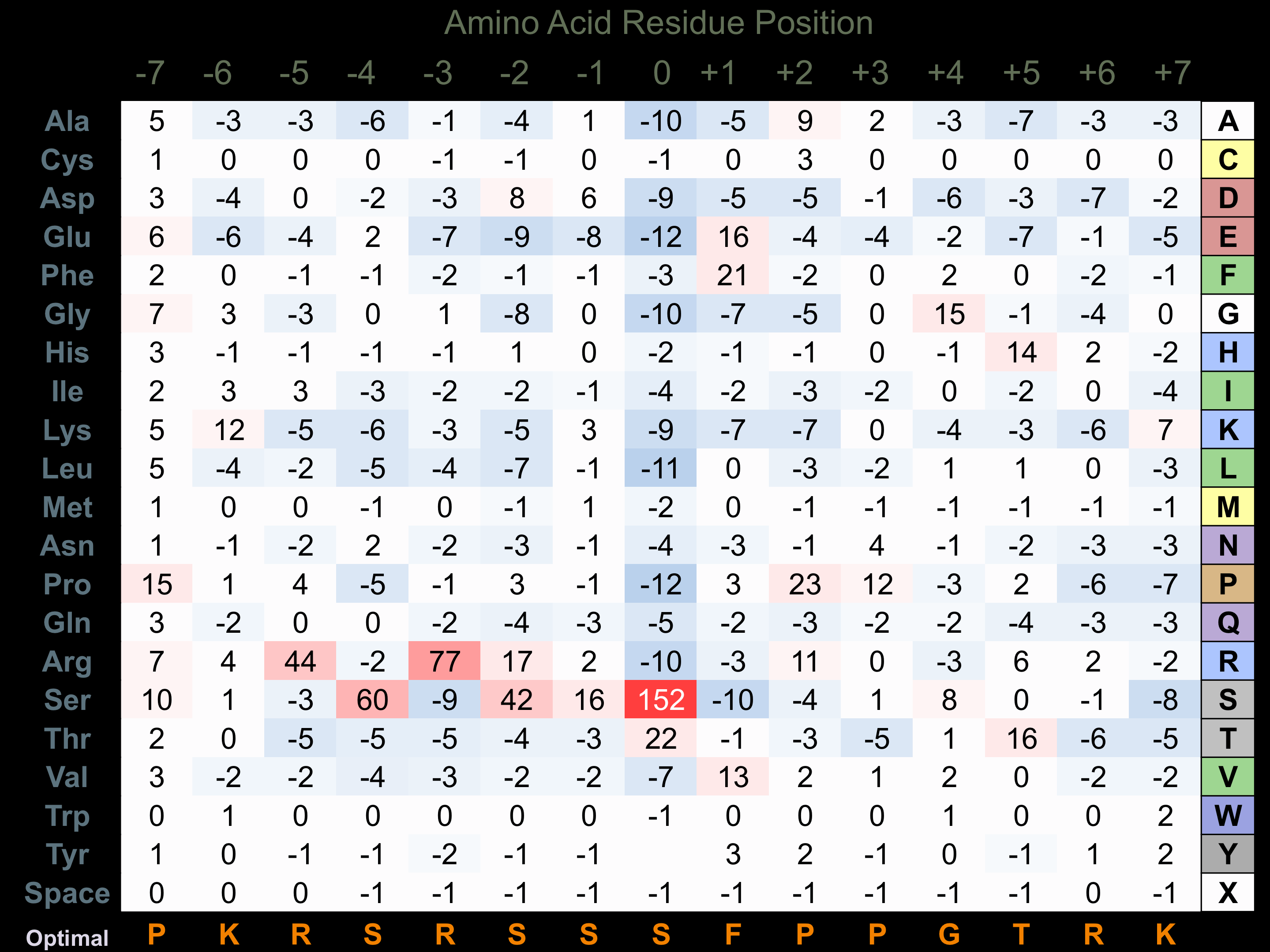
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Sorafenib | IC50 > 50 nM | 216239 | 1336 | 22037377 |
| Dasatinib | IC50 > 250 nM | 11153014 | 1421 | 22037377 |
| PD169316 | IC50 > 250 nM | 4712 | 17331 | 22037377 |
| PKR Inhibitor; Negative Control | IC50 > 250 nM | 16760619 | 22037377 | |
| SB202190 | IC50 > 250 nM | 5353940 | 278041 | 22037377 |
| SB203580 | IC50 > 250 nM | 176155 | 10 | 22037377 |
| Lck Inhibitor | IC50 = 500 nM | 6603792 | 22037377 | |
| p38 MAP Kinase Inhibitor | IC50 = 500 nM | 4665 | 91730 | 22037377 |
| 4557W | IC50 > 1 µM | 9843206 | 563845 | 22037377 |
| BML-275 | IC50 > 1 µM | 11524144 | 478629 | 22037377 |
| TWS119 | IC50 > 1 µM | 9549289 | 405759 | 22037377 |
Disease Linkage
General Disease Association:
Cancer, bacterial infection
Specific Diseases (Non-cancerous):
Mycobacterium marinum infection
Comments:
Relatively rare, Infection by mycobacterium marinum is similar to arthritis and tuberculosis in its symptoms. Affected tissues include Affiliated tissues include skin, testes and lymph node, and its also affects the immune and hematopoietic systems.
Specific Cancer Types:
Colorectal adenocarcinoma
Comments:
A-RAF may be an oncoprotein (OP) based on its similarity to Raf1 and B-Raf. However, it does not display a higher than normal rate of mutation in human tumours. The active form of the protein kinase normally acts to promote tumour cell proliferation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -92, p<0.0001); Brain oligodendrogliomas (%CFC= -67, p<0.0001); Ovary adenocarcinomas (%CFC= +54, p<0.044); and Prostate cancer - primary (%CFC= -98, p<0.0001). The COSMIC website notes an up-regulated expression score for A-RAF in diverse human cancers of 606, which is 1.3-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 172 for this protein kinase in human cancers was 2.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25612 diverse cancer specimens. This rate is very similar (+ 2% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.38 % in 602 endometrium cancers tested; 0.37 % in 1208 large intestine cancers tested; 0.34 % in 589 stomach cancers tested; 0.13 % in 1942 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S214F (3).
Comments:
Only 9 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.

