Nomenclature
Short Name:
SRC
Full Name:
Proto-oncogene tyrosine-protein kinase Src
Alias:
- ASV
- C-Src
- EC 2.7.10.2
- P60-SRC
- P60-Src
- SRC1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Src
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
59835
# Amino Acids:
536
# mRNA Isoforms:
2
mRNA Isoforms:
60,589 Da (542 AA; P12931-2); 59,835 Da (536 AA; P12931)
4D Structure:
Interacts with DDEF1/ASAP1; via the SH3 domain. Interacts with CCPG1 By similarity. Interacts with CDCP1, PELP1, TGFB1I1 and TOM1L2. Interacts with the cytoplasmic domain of MUC1, phosphorylates it and increases binding of MUC1 with beta-catenin. Interacts with RALGPS1; via the SH3 domain. Interacts with HEV ORF3 protein; via the SH3 domain. Interacts with CAV2 (tyrosine phosphorylated form). Interacts (via the SH3 domain and the protein kinase domain) with ARRB1; the interaction is independent of the phosphorylation state of SRC C-terminus. Interacts with ARRB1 and ARRB2. Interacts with SRCIN1. Interacts with NDFIP2 and more weakly with NDFIP1
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Threonine phosphorylated:
T37, T72, T74, T182, T183, T246, T250, T304, T420+, T526.
Tyrosine phosphorylated:
Y93, Y139, Y187, Y216+, Y338, Y385, Y419+, Y439, Y522, Y530-.
Ubiquitinated:
K430.
Acetylated:
K62.
Myristoylated:
G2.
Serine phosphorylated:
S12+, S17-, S35, S39, S43, S51, S69, S70, S75+, S97-, S104, S269, S306, S348, S525.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 91
91
823
69
1039
 3
3
26
25
36
 58
58
525
13
556
 23
23
211
220
600
 57
57
516
62
393
 3
3
24
152
69
 16
16
144
87
401
 96
96
868
50
1495
 44
44
399
24
360
 11
11
101
123
176
 8
8
76
39
121
 74
74
672
278
606
 9
9
86
46
167
 1.4
1.4
13
14
10
 27
27
244
33
560
 5
5
49
43
79
 7
7
62
229
139
 13
13
122
23
175
 4
4
38
134
78
 40
40
361
254
336
 11
11
101
28
126
 7
7
61
36
97
 33
33
303
24
497
 16
16
145
25
228
 10
10
94
29
138
 81
81
740
141
1224
 5
5
47
49
98
 19
19
171
24
217
 26
26
236
25
374
 8
8
69
42
30
 49
49
444
42
399
 100
100
908
77
1690
 13
13
115
124
453
 78
78
706
187
619
 6
6
57
118
58
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 92.7
92.7
95.3
100 99.8
99.8
99.8
100 -
-
-
98 -
-
-
- 98.9
98.9
98.9
99 -
-
-
- 97.8
97.8
98.3
99 98.5
98.5
99.3
99 -
-
-
- 70.9
70.9
78.7
- 94.4
94.4
96.3
95 87.7
87.7
92.2
89 83.2
83.2
88.8
85 -
-
-
- 54.1
54.1
67.2
53 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
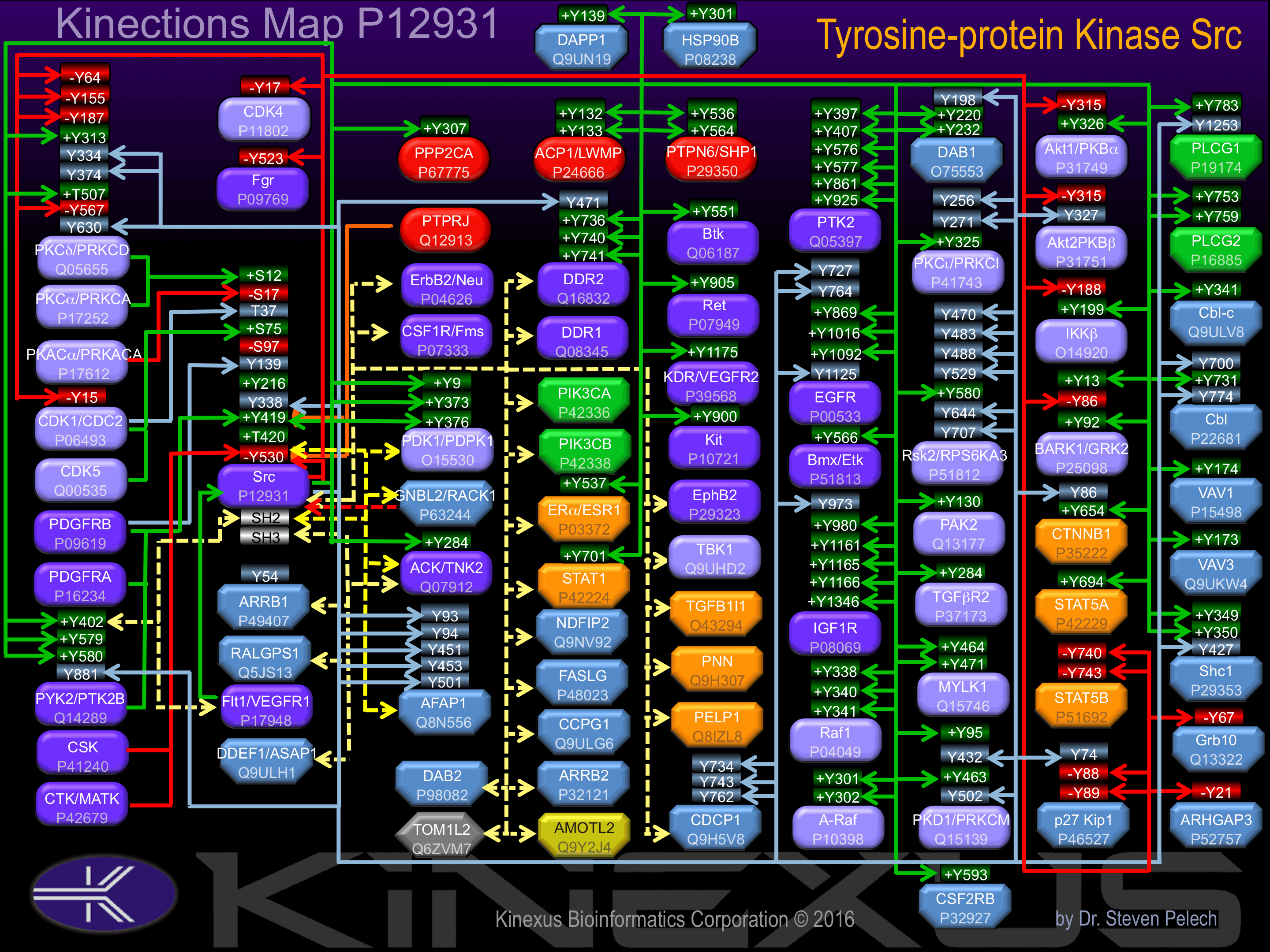
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BCAR1 - P56945 |
| 2 | ESR1 - P03372 |
| 3 | EGFR - P00533 |
| 4 | CSK - P41240 |
| 5 | CBL - P22681 |
| 6 | DAG1 - Q14118 |
| 7 | RAF1 - P04049 |
| 8 | STAT3 - P40763 |
| 9 | ADRB2 - P07550 |
| 10 | PIK3R1 - P27986 |
| 11 | PXN - P49023 |
| 12 | CDCP1 - Q9H5V8 |
| 13 | ERBB2 - P04626 |
| 14 | LRP1 - Q07954 |
| 15 | PELP1 - Q8IZL8 |
Regulation
Activation:
Phosphorylation of Ser-12 increases phosphotransferase activity and interaction with PPP2R2C. Phosphorylation of Ser-75 and Thr-420 increases phosphotransferase activity. Phosphorylation of Tyr-419 increases phosphotransferase activity and induces interaction with Cbl-c.
Inhibition:
Phosphorylation at S17 may reduce phosphorylation at S12. Phosphorylation of Ser-97 and Tyr-530 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKCa | P17252 | S12 | KSKPKDASQRRRSLE | + |
| PKCd | Q05655 | S12 | KSKPKDASQRRRSLE | + |
| PKACa | P17612 | S17 | DASQRRRSLEPAENV | - |
| CDK1 | P06493 | T37 | GAFPASQTPSKPASA | |
| CDK1 | P06493 | S75 | NSSDTVTSPQRAGPL | + |
| CDK5 | Q00535 | S75 | NSSDTVTSPQRAGPL | + |
| PDGFRB | P09619 | Y139 | TGYIPSNYVAPSDSI | |
| SRC | P12931 | Y338 | VVSEEPIYIVTEYMS | |
| PKACa | P17612 | Y419 | RLIEDNEYTARQGAK | + |
| PDGFRB | P09619 | Y419 | RLIEDNEYTARQGAK | + |
| PDGFRA | P16234 | Y419 | RLIEDNEYTARQGAK | + |
| PYK2 | Q14289 | Y419 | RLIEDNEYTARQGAK | + |
| SRC | P12931 | Y419 | RLIEDNEYTARQGAK | + |
| FAK | Q05397 | Y419 | RLIEDNEYTARQGAK | + |
| CSK | P41240 | Y530 | FTSTEPQYQPGENL_ | - |
| SRC | P12931 | Y530 | FTSTEPQYQPGENL_ | - |
| CTK | P42679 | Y530 | FTSTEPQYQPGENL_ | - |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| p27Kip1 | P46527 | Y88 | KGSLPEFYYRPPRPP | - |
| p27Kip1 | P46527 | Y89 | GSLPEFYYRPPRPPK | - |
| p70S6K (RPS6KB1) | P23443 | S375 | PFKPLLQSEEDVSQF | |
| p70S6K (RPS6KB1) | P23443 | S380 | LQSEEDVSQFDSKFT | |
| p70S6K (RPS6KB1) | P23443 | T250 | SIHDGTVTHTFCGTI | + |
| p70S6K (RPS6KB1) | P23443 | Y62 | DHGGVGPYELGMEHC | |
| p70S6Kb (RPS6KB2) | Q9UBS0 | Y45 | GLEPVGHYEEVELTE | |
| PAG | Q9NWQ8 | Y317 | EEEISAMYSSVNKPG | |
| PAK2 | Q13177 | Y130 | VLDVLKFYDSNTVKQ | + |
| PDK1 (PDPK1) | O15530 | Y373 | SEDDEDCYGNYDNLL | + |
| PDK1 (PDPK1) | O15530 | Y376 | DEDCYGNYDNLLSQF | + |
| PDK1 (PDPK1) | O15530 | Y9 | ARTTSQLYDAVPIQS | + |
| PELP1 | Q8IZL8 | Y920 | EEEDEEEYFEEEEEE | ? |
| PFN1 | P07737 | Y129 | GLINkkCYEMASHLR | |
| PIP5KG | O60331 | Y649 | TDERSWVYSPLHYSA | |
| PKCd (PRKCD) | Q05655 | T507 | FGESRASTFCGTPDY | + |
| PKCd (PRKCD) | Q05655 | Y155 | IKQAKIHYIKNHEFI | - |
| PKCd (PRKCD) | Q05655 | Y187 | WGLNKQGYKCRQCNA | - |
| PKCd (PRKCD) | Q05655 | Y313 | SSEPVGIYQGFEKKT | + |
| PKCd (PRKCD) | Q05655 | Y334 | MQDNSGTYGKIWEGS | ? |
| PKCd (PRKCD) | Q05655 | Y374 | ELKGRGEYSAIKALK | |
| PKCd (PRKCD) | Q05655 | Y567 | IRVDTPHYPRWITKE | - |
| PKCd (PRKCD) | Q05655 | Y630 | KVKSPRDYSNFDQEF | |
| PKCd (PRKCD) | Q05655 | Y64 | STFDAHIYEGRVIQI | - |
| PKCi (PRKCI) | P41743 | Y256 | RVIGRGSYAKVLLVR | |
| PKCi (PRKCI) | P41743 | Y271 | LKKTDRIYAMKVVKK | |
| PKCi (PRKCI) | P41743 | Y325 | RLFFVIEYVNGGDLM | + |
| PKD1 (PRKCM) | Q15139 | Y432 | KEGWMVHYTSKDTLR | |
| PKD1 (PRKCM) | Q15139 | Y463 | NDTGSRYYKEIPLSE | + |
| PKD1 (PRKCM) | Q15139 | Y502 | TTANVVYYVGENVVN | |
| PKD1 (PRKCM) | Q15139 | Y95 | KFPECGFYGMYDKIL | + |
| PLCG1 | P19174 | Y1253 | EGSFESRYQQPFEDF | |
| PLCG1 | P19174 | Y783 | EGRNPGFYVEANPMP | + |
| PLCG2 | P16885 | Y753 | ERDINSLYDVSRMYV | + |
| PLCG2 | P16885 | Y759 | LYDVSRMYVDPSEIN | + |
| PLSCR1 | O15162 | Y69 | PVPNQPVYNQPVYNQ | |
| PLSCR1 | O15162 | Y74 | PVYNQPVYNQPVGAA | |
| Polycystin 1 | P98161 | Y4237 | NQATEDVYQLEQQLH | |
| PPP2CA | P67775 | Y307 | VTRRTPDYFL_____ | + |
| PROM1 | O43490 | Y828 | RMDSEDVYDDVETIP | |
| PROM1 | O43490 | Y852 | GYHKDHVYGIHNPVM | |
| PSF | P23246 | Y597 | RRQREESYSRMGYMD | |
| PSF | P23246 | Y691 | GPGTPAGYGRGREEY | |
| PTEN | P60484 | Y240 | RREDKFMYFEFPQPL | + |
| PTPN6 (SHP1) | P29350 | Y536 | QKGQESEYGNITYPP | + |
| PTPN6 (SHP1) | P29350 | Y564 | SKHKEDVYENLHTKN | + |
| PTPRA | P18433 | Y798 | YIDAFSDYANFK___ | ? |
| PXN | P49023 | Y118 | VGEEEHVYSFPNKQK | |
| PXN | P49023 | Y31 | FLSEETPYSYPTGNH | |
| PXN | P49023 | Y40 | YPTGNHTYQEIAVPP | |
| PXN | P49023 | Y88 | PQSSSPVYGSSAKTS | |
| PYK2 (PTK2B) | Q14289 | Y402 | CSIESDIYAEIPDET | + |
| PYK2 (PTK2B) | Q14289 | Y579 | RYIEDEDYYKASVTR | + |
| PYK2 (PTK2B) | Q14289 | Y580 | YIEDEDYYKASVTRL | + |
| PYK2 (PTK2B) | Q14289 | Y881 | DRTDDLVYLNVMELV | |
| R-Ras | P10301 | Y66 | DPTIEDSYTKICSVD | |
| RACK1 | P63244 | Y228 | LNEGKHLYTLDGGDI | |
| RACK1 | P63244 | Y246 | LCFSPNRYWLCAATG | |
| Raf1 | P04049 | S338 | RPRGQRDSSYYWEIE | + |
| Raf1 | P04049 | Y340 | RGQRDSSYYWEIEAS | + |
| Raf1 | P04049 | Y341 | GQRDSSYYWEIEASE | + |
| RapGEF1 | Q13905 | Y504 | APIPSVPYAPFAAIL | |
| RBM10 | P98175 | Y16 | RGDRTGRYGATDRSQ | |
| RBM10 | P98175 | Y434 | SEEPPVDYSYYQQDE | |
| RBM10 | P98175 | Y905 | LGARGSSYGVTSTES | |
| Ret | P07949 | Y905 | DVYEEDSYVKRSQGR | + |
| RGS16 | O15492 | Y168 | TLMEKDSYPRFLKSP | |
| RGS16 | O15492 | Y177 | RFLKSPAYRDLAAQA | |
| RHBG | Q9H310-6 | Y429 | SPPDSQHYEDQVHWQ | |
| ROMK | P48048 | Y337 | SKTKEGKYRVDFHNF | |
| RSK2 (RPS6KA3) | P51812 | Y470 | EIEILLRYGQHPNII | |
| RSK2 (RPS6KA3) | P51812 | Y483 | IITLKDVYDDGKYVY | |
| RSK2 (RPS6KA3) | P51812 | Y488 | DVYDDGKYVYVVTEL | |
| RSK2 (RPS6KA3) | P51812 | Y529 | TITKTVEYLHAQGVV | ? |
| RSK2 (RPS6KA3) | P51812 | Y580 | GLLMTPCYTANFVAP | + |
| RSK2 (RPS6KA3) | P51812 | Y644 | KFSLSGGYWNSVSDT | |
| RSK2 (RPS6KA3) | P51812 | Y707 | KGAMAATYSALNRNQ | |
| SF3B2 | Q13435 | Y356 | VTEEPEIYEPNFIFF | |
| SF3B2 | Q13435 | Y569 | TIHGDLYYEGKEFET | |
| SH3GL1 | Q99961 | Y315 | QPSCKALYDFEPEND | |
| SH3GLB1 | Q9Y371 | Y80 | ARIEEFVYEKLDRKA | |
| Shc1 | P29353 | Y349 | EEPPDHQYYNDFPGK | + |
| Shc1 | P29353 | Y350 | EPPDHQYYNDFPGKE | + |
| Shc1 | P29353 | Y427 | ELFDDPSYVNVQNLD | ? |
| Shc3 | Q92529 | Y341 | GDGSDHPYYNSIPSK | ? |
| Shc3 | Q92529 | Y342 | DGSDHPYYNSIPSKM | ? |
| Shc3 | Q92529 | Y379 | FAGKEQTYYQGRHLG | |
| Shc3 | Q92529 | Y380 | AGKEQTYYQGRHLGD | |
| Shc3 | P29353 | Y406 | RQGSSDIYSTPEGKL | |
| Shc3 | P29353 | Y424 | PTGEAPTYVNTQQIP | |
| SNCA | P37840 | Y125 | VDPDNEAYEMPSEEG | |
| Spry2 | O43597 | Y55 | AIRNTNEYTEGPTVV | |
| SPTAN1 | Q13813 | Y1073 | MKQVEELYHSLLELG | |
| SPTAN1 | Q13813 | Y1176 | AVQQQEVYGMMPRDE | |
| Src | P12931 | Y338 | VVSEEPIYIVTEYMS | |
| Src | P12931 | Y419 | RLIEDNEYTARQGAK | + |
| Src | P12931 | Y530 | FTSTEPQYQPGENL_ | - |
| STAT1 | P42224 | Y701 | DGPKGTGYIKTELIS | + |
| STAT5A | P42229 | Y694 | LAKAVDGYVKPQIKQ | + |
| STAT5B | P51692 | Y740 | AVCPQAHYNMYPQNP | - |
| STAT5B | P51692 | Y743 | PQAHYNMYPQNPDSV | - |
| SYN1 | P17600 | Y301 | VVALTKTYATAEPFI | |
| Syndecan-2 | P34741 | Y179 | RKKDEGSYDLGERKP | |
| Tau iso8 | P10636-8 | Y18 | MEDHAGTYGLGDRKD | |
| TERT | O14746 | Y707 | QDPPPELYFVKVDVT | |
| TGFbR2 | P37173 | Y284 | KIFPYEEYASWKTEK | + |
| THRAP3 | Q9Y2W1 | Y107 | NRGGYGNYRSNWQNY | |
| THRAP3 | Q9Y2W1 | Y118 | WQNYRQAYSPRRGRS | |
| THRAP3 | Q9Y2W1 | Y344 | AASGGAAYTKRYLEE | |
| THRAP3 | Q9Y2W1 | Y68 | ERNHPRVYQNRDFRG | |
| Tiam1 | Q13009 | Y384 | DAARQGVYENFRREL | |
| TRIP6 | Q15654 | Y55 | PLPSEQCYQAPGGPE | |
| TRPC3 | Q13507 | Y148 | ELQDDDFYAYDEDGT | |
| TRPC3 | Q13507 | Y150 | QDDDFYAYDEDGTRF | |
| TRPC3 | Q13507 | Y226 | KGLASPAYLSLSSED | |
| TRPC3 | Q13507 | Y49 | RFLDAAEYGNIPVVR | |
| TRPV4 | Q9HBA0 | Y110 | PMDSLFDYGTYRHHS | |
| TRPV4 | Q9HBA0 | Y805 | DPGKNETYQYYGFSH | |
| TRPV6 | Q9H1D0 | Y161 | RSPCNLIYFGEHPLS | |
| UGT2B7 | P16662 | Y438 | RVINDPSYKENVMKL | |
| VAV1 | P15498 | Y174 | EAEGDEIYEDLMRSE | + |
| VAV3 | Q9UKW4 | Y173 | EDEGGEVYEDLMKAE | + |
| Villin | P09327 | Y255 | LKAALKLYHVSDSEG | |
| Villin | P09327 | Y45 | SFFDGDCYIILAIHK | |
| Villin | P09327 | Y59 | KTASSLSYDIHYWIG | |
| Villin | P09327 | Y63 | SLSYDIHYWIGQDSS | |
| Villin | P09327 | Y80 | EQGAAAIYTTQMDDF | |
| Vinculin | P18206 | Y1132 | WVRKTPWYQ______ | |
| Vinculin | P18206 | Y99 | QMLQSDPYSVPARDY | |
| VR1 | Q8NER1 | Y200 | ASYTDSYYKGQTALH | |
| WASF1 | Q92558 | Y125 | PIPLQETYDVCEQPP | |
| WWOX | Q9NZC7 | Y33 | TTKDGWVYYANHTEE | |
| ZC3H4 | Q9UPT8 | Y1218 | PTDRYNSYNRPRPKA | |
| ZC3H4 | Q9UPT8 | Y158 | GHRKYREYSPPYAPS | |
| ZC3H4 | Q9UPT8 | Y247 | SRGRGRGYRGRGSRG | |
| 14-3-3 zeta (YWHAZ) | P63104 | Y179 | LNFSVFYYEILNSPE | |
| A-Raf | P10398 | Y301 | LGYRDSGYYWEVPPS | + |
| A-Raf | P10398 | Y302 | GYRDSGYYWEVPPSE | + |
| A2LP | Q8WWM7 | Y744 | VPGQQGKYRGAKGSL | |
| ACK1 (TNK2) | Q07912 | Y284 | LPQNDDHYVMQEHRK | + |
| ACP1 (Low Mr PTPase) | P24666 | Y132 | QLIIEDPYYGNDSDF | + |
| ACP1 (Low Mr PTPase) | P24666 | Y133 | LIIEDPYYGNDSDFE | + |
| ADAM12 | O43184 | Y907 | PRSTHTAYIK_____ | |
| AFAP | Q8N556 | Y451 | TDPEALHYDYIDVEM | |
| AFAP | Q8N556 | Y453 | PEALHYDYIDVEMSA | |
| AFAP | Q8N556 | Y501 | PSGTALHYDDVPCIN | |
| AFAP | Q8N556 | Y93 | TSSLPEGYYEEAVPL | |
| AFAP | Q8N556 | Y94 | SSLPEGYYEEAVPLS | |
| AIP1 | Q86UL8 | Y319 | YTEKGEVYFIDHNTK | |
| AKAP8 | Q8NE02 | Y152 | PYRPSYSYDYEFDLG | |
| Akt1 (PKBa) | P31749 | Y315 | TFCGTPEYLAPEVLE | - |
| Akt1 (PKBa) | P31749 | Y326 | EVLEDNDYGRAVDWW | + |
| Akt2 (PKBb) | P31751 | Y316 | TFCGTPEYLAPEVLE | - |
| Akt2 (PKBb) | P31751 | Y327 | EVLEDNDYGRAVDWW | |
| ANXA1 | P04083 | Y20 | IENEEQEYVQTVKSS | |
| ANXA2 | P07355 | Y23 | HSTPPSAYGSVKAYT | |
| AP2B1 | P63010 | Y737 | THRQGHIYMEMNFTN | |
| APP | P05067 | Y757 | SKMQQNGYENPTYKF | |
| ARAP3 | Q8WWN8 | Y1403 | EELEEPVYEEPVYEE | |
| ARAP3 | Q8WWN8 | Y1408 | PVYEEPVYEEVGAFP | |
| ARHGAP3 | P52757 | Y21 | VSSDAEEYQPPIWKS | - |
| ARHGAP5 | Q13017 | Y1091 | DMDPSDNYAEPIDTI | |
| ARHGAP5 | Q13017 | Y1109 | KGYSDEIYVVPDDSQ | |
| ARHGDIA | P52565 | Y156 | YGPRAEEYEFLTPVE | |
| ARHGDIA | P52565 | Y27 | EDEHSVNYKPPAQKS | |
| ARHGDIB | P52566 | Y153 | YGPRPEEYEFLTPVE | |
| ARHGDIB | P52566 | Y24 | ELDSKLNYKPPPQKS | ? |
| ARHGEF4 iso3 | Q9NR80-3 | Y94 | VGSEEDLYDDLHSSS | |
| ARRB1 | P49407 | Y54 | YLKERRVYVTLTCAF | |
| ARS2 | Q9BXP5 | Y175 | AVKRYNDYKLDFRRQ | |
| ARS2 | Q9BXP5 | Y798 | TPQGLMPYGQPRPPI | |
| ARS2 | Q9BXP5 | Y836 | YGAGRGNYDAFRGQG | |
| ARS2 | Q9BXP5 | Y92 | DEHSSDPYHSGYEMP | |
| AT1 (Angiotensin II receptor) | P30556 | Y319 | YFLQLLKYIPPKAKS | |
| BARK1 (GRK2, ADRBK1) | P25098 | Y13 | AVLADVSYLMAMEKS | + |
| BARK1 (GRK2, ADRBK1) | P25098 | Y86 | ARPLVEFYEEIKKYE | - |
| BARK1 (GRK2, ADRBK1) | P25098 | Y92 | FYEEIKKYEKLETEE | + |
| BCLAF1 | Q9NYF8 | Y150 | SSRSSSPYSKSPVSK | |
| BCLAF1 | Q9NYF8 | Y284 | VGNGSSRYSPSQNSP | |
| BCLAF1 | Q9NYF8 | Y408 | DTEETEDYRQFRKSV | |
| BCLAF1 | Q9NYF8 | Y80 | YRGRGRGYYQGGGGR | |
| BCLAF1 | Q9NYF8 | Y81 | RGRGRGYYQGGGGRY | |
| BCLAF1 | Q9NYF8 | Y93 | GRYHRGGYRPVWNRR | |
| BKR2 | P30411 | Y177 | GVRWAKLYSLVIWGC | |
| BKR2 | P30411 | Y347 | RKKSWEVYQGVCQKG | |
| Bmx (Etk) | P51813 | Y566 | RYVLDDQYVSSVGTK | + |
| Btk | Q06187 | Y551 | RYVLDDEYTSSVGSK | + |
| CACNA1B | Q00975 | Y788 | RASCEALYSEMDPEE | |
| CACNA1C | Q13936 | Y2217 | ELQDSRVYVSSL___ | |
| Calmodulin | P62158 | Y139 | DGDGQVNYEEFVQMM | |
| Calmodulin | P62158 | Y99 | FDKDGNGYISAAELR | |
| Calponin 2 | Q99439 | Y11 | QFNKGPSYGLSAEVK | |
| Calponin 3 | Q15417 | Y10 | HFNKGPSYGLSAEVK | |
| Caspase 8 | Q14790 | Y380 | TDSEEQPYLEMDLSS | |
| Caveolin 1 (CAV1) | Q03135 | Y14 | VDSEGHLYTVPIREQ | |
| Caveolin 2 (CAV2) | P51636 | Y19 | LFMDDDSYSHHSGLE | |
| Cbl | P22681 | Y700 | EGEEDTEYMTPSSRP | |
| Cbl | P22681 | Y731 | QQIDSCTYEAMYNIQ | + |
| Cbl | P22681 | Y774 | SENEDDGYDVPKPPV | |
| Cbl-c | Q9ULV8 | Y341 | SEEQLQLYWAMDSTF | + |
| CD46 iso4 | P15529-4 | Y354 | KADGGAEYATYQTKS | |
| Cdc42 | P60953 | Y64 | DTAGQEDYDRLRPLS | |
| CDCP1 | Q9H5V8 | Y734 | KDNDSHVYAVIEDTM | |
| CDCP1 | Q9H5V8 | Y743 | VIEDTMVYGHLLQDS | |
| CDCP1 | Q9H5V8 | Y762 | LQPEVDTYRPFQGTM | |
| CDH2 | P19022 | Y820 | PIHAEPQYPVRSAAP | |
| CDH2 | P19022 | Y852 | NDPTAPPYDSLLVFD | |
| CDH2 | P19022 | Y860 | DSLLVFDYEGSGSTA | |
| CDH2 | P19022 | Y884 | SSGGEQDYDYLNDWG | |
| CDH2 | P19022 | Y886 | GGEQDYDYLNDWGPR | |
| CDH5 | P33151 | Y658 | GEMDTTSYDVSVLNS | |
| CDH5 | P33151 | Y685 | LDARPSLYAQVQKPP | |
| CDH5 | P33151 | Y731 | PYDTLHIYGYEGSES | |
| CDK1 (CDC2) | P06493 | Y15 | EKIGEGTYGVVYKGR | - |
| CDK4 | P11802 | Y17 | AEIGVGAYGTVYKAR | - |
| CLTC | Q00610 | Y1477 | LFITEEDYQALRTSI | |
| CLTCL1 | P53675 | Y1477 | LLTEEEDYQGLRASI | |
| Coronin 7 | P57737 | Y288 | GERQLYCYEVVPQQP | |
| Coronin 7 | P57737 | Y758 | GDTRVFLYELLPESP | |
| Cortactin | Q14247 | Y334 | VTQVSSAYQKTVPVE | |
| Cortactin | Q14247 | Y421 | RLPSSPVYEDAASFK | |
| Cortactin | Q14247 | Y470 | AYATEAVYESAEAPG | |
| Cortactin | Q14247 | Y486 | YPAEDSTYDEYENDL | |
| CSF2RB | P32927 | Y593 | SFDFNGPYLGPPHSR | + |
| CTNNB1 | P35222 | Y654 | RNEGVATYAAAVLFR | + |
| CTNNB1 | P35222 | Y86 | VADIDGQYAMTRAQR | |
| CTNND1 | O60716 | Y112 | PGQIVETYTEEDPEG | |
| CTNND1 | O60716 | Y217 | PDGYSRHYEDGYPGG | |
| CTNND1 | O60716 | Y228 | YPGGSDNYGSLSRVT | |
| CTNND1 | O60716 | Y257 | APSRQDVYGPQPQVR | |
| CTNND1 | O60716 | Y280 | HRFHPEPYGLEDDQR | |
| CTNND1 | O60716 | Y291 | DDQRSMGYDDLDYGM | |
| CTNND1 | O60716 | Y296 | MGYDDLDYGMMSDYG | |
| CTNND1 | O60716 | Y302 | DYGMMSDYGTARRTG | |
| CTNND1 | O60716 | Y96 | QDHSHLLYSTIPRMQ | |
| Cx43 | P17302 | Y246 | VKGRSDPYHATTGPL | |
| Cx43 | P17302 | Y264 | KDCGSQKYAYFNGCS | |
| DAB1 | O75553 | Y198 | EDVEDPVYQYIVFEA | |
| DAB1 | O75553 | Y220 | PETEENIYQVPTSQK | + |
| DAB1 | O75553 | Y232 | SQKKEGVYDVPKSQP | + |
| DAG1 | Q14118 | Y892 | PYRSPPPYVPP____ | |
| DAPP1 | Q9UN19 | Y139 | KVEEPSIYESVRVHT | + |
| DDR2 | Q16832 | Y471 | EQGSNSTYDRIFPLR | |
| DDR2 | Q16832 | Y736 | FGMSRNLYSGDYYRI | + |
| DDR2 | Q16832 | Y740 | RNLYSGDYYRIQGRA | + |
| DDR2 | Q16832 | Y741 | NLYSGDYYRIQGRAV | + |
| DGK-A | P23743 | Y335 | ILPPSSIYPSVLASG | |
| DLG4 (PSD-95) | P78352 | Y523 | REDSVLSYETVTQME | |
| DYN1 | Q05193 | Y231 | LLPLRRGYIGVVNRS | |
| DYN1 | Q05193 | Y597 | NTEQRNVYKDYRQLE | |
| DYN2 | P50570 | Y231 | LLPLRRGYIGVVNRS | |
| DYN2 | P50570 | Y597 | NTEQRNVYKDLRQIE | |
| EGFR | P00533 | Y1016 | DVVDADEYLIPQQGF | + |
| EGFR | P00533 | Y1092 | TFLPVPEYINQSVPK | + |
| EGFR | P00533 | Y1125 | APSRDPHYQDPHSTA | |
| EGFR | P00533 | Y727 | SGAFGTVYKGLWIPE | |
| EGFR | P00533 | Y764 | KEILDEAYVMASVDN | |
| EGFR | P00533 | Y869 | LGAEEKEYHAEGGKV | + |
| eNOS | P29474 | Y81 | WEVGSITYDTLSAQA | |
| Ephexin1 | Q8N5V2 | Y179 | IEQIGLLYQEYRDKS | |
| Ephrin-B1 | P98172 | Y324 | YEKVSGDYGHPVYIV | |
| Ephrin-B1 | P98172 | Y329 | GDYGHPVYIVQEMPP | |
| ERa (ESR1) | P03372 | Y537 | CKNVVPLYDLLLEML | + |
| EWS | Q01844 | Y278 | HPSSMGVYGQESGGF | |
| Ezrin | P15311 | Y146 | KEVHKSGYLSSERLI | |
| Ezrin | P15311 | Y478 | PPPPPPVYEPVSYHV | |
| FAK (PTK2) | Q05397 | Y397 | SVSETDDYAEIIDEE | + |
| FAK (PTK2) | Q05397 | Y407 | IIDEEDTYTMPSTRD | + |
| FAK (PTK2) | Q05397 | Y576 | RYMEDSTYYKASKGK | + |
| FAK (PTK2) | Q05397 | Y577 | YMEDSTYYKASKGKL | + |
| FAK (PTK2) | Q05397 | Y861 | PIGNQHIYQPVGKPD | + |
| FAK (PTK2) | Q05397 | Y925 | DRSNDKVYENVTGLV | + |
| FBP1 | Q96AE4 | Y268 | FREVRNEYGSRIGGN | |
| FBP1 | Q96AE4 | Y58 | TSLNSNDYGYGGQKR | |
| FBP1 | Q96AE4 | Y625 | YYRQQAAYYAQTSPQ | |
| FcRL3 | Q96P31 | Y650 | PMELEPMYSNVNPGD | |
| FcRL3 | Q96P31 | Y662 | PGDSNPIYSQIWSIQ | |
| FcRL3 | Q96P31 | Y692 | HEELTVLYSELKKTH | |
| FcRL3 | Q96P31 | Y722 | EEDDEENYENVPRVL | |
| Fgr | P09769 | Y523 | FTSAEPQYQPGDQT_ | - |
| FHIT | P49789 | Y113 | FHRNDSIYEELQKHD | |
| FIP1L1 | Q6UN15 | Y110 | IKTGAPQYGSYGTAP | |
| FLOT2 | Q14254 | Y114 | DVYDKVDYLSSLGKT | |
| G3BP1 | Q13283 | Y133 | VHNDIFRYQDEVFGG | |
| G6PD | P11413 | Y428 | DLTYGNRYKNVKLPD | |
| G6PD | P11413 | Y507 | GFQYEGTYKWVNPHK | |
| GABRG2 | P18507 | Y404 | LQERDEEYGYECLDG | |
| GABRG2 | P18507 | Y406 | ERDEEYGYECLDGKD | |
| Gelsolin | P06396 | Y409 | TDGLGLSYLSSHIAN | |
| Gelsolin | P06396 | Y465 | VPVDPATYGQFYGGD | |
| Gelsolin | P06396 | Y603 | LKTPSAAYLWVGTGA | |
| Gelsolin | P06396 | Y651 | ALGGKAAYRTSPRLK | |
| Gelsolin | P06396 | Y86 | VPVPTNLYGDFFTGD | |
| GLRB | P48167 | Y435 | RDFELSNYDCYGKPI | |
| GNAT1 | P11488 | Y141 | CFERASEYQLNDSAG | |
| Grb10 | Q13322 | Y67 | NASLESLYSACSMQS | - |
| Grb2 | P62993 | Y160 | QVPQQPTYVQALFDF | |
| GRF1 | Q9NRY4 | Y1105 | RNEEENIYSVPHDST | |
| GTF2I | P78347 | Y248 | EESEDPDYYQYNIQG | |
| GTF2I | P78347 | Y652 | KPELVISYLPPGMAS | |
| GUCY1B3 | Q02153 | Y192 | ESKEEDFYEDLDRFE | |
| Hcn2 | Q9UL51 | Y503 | FRQKIHDYYEHRYQG | |
| HCN4 | Q9Y3Q4 | Y531 | RRQYQEKYKQVEQYM | |
| HCRTR1 | O43613 | Y358 | SAANPIIYNFLSGKF | |
| hnRNP K | P61978 | Y225 | IKGRAQPYDPNFYDE | |
| hnRNP K | P61978 | Y230 | QPYDPNFYDETYDYG | |
| hnRNP K | P61978 | Y234 | PNFYDETYDYGGFTM | |
| hnRNP K | P61978 | Y236 | FYDETYDYGGFTMMF | |
| hnRNP K | P61978 | Y380 | YAGGRGSYGDLGGPI | |
| hnRNP K | P61978 | Y458 | LQNSVKQYSGKFF__ | |
| hnRNP K | P61978 | Y72 | IKALRTDYNASVSVP | |
| HSP90B | P08238 | Y301 | DDITQEEYGEFYKSL | + |
| IGF1R | P08069 | Y1161 | FGMTRDIYETDYYRK | + |
| IGF1R | P08069 | Y1165 | RDIYETDYYRKGGKG | + |
| IGF1R | P08069 | Y1166 | DIYETDYYRKGGKGL | + |
| IGF1R | P08069 | Y1346 | SFDERQPYAHMNGGR | + |
| IGF1R | P08069 | Y973 | RLGNGVLYASVNPEY | |
| IGF1R | P08069 | Y980 | YASVNPEYFSAADVY | + |
| IkBa | P25963 | Y42 | DSMKDEEYEQMVKEL | |
| IKKb (IKBKINASE) | O14920 | Y188 | SFVGTLQYLAPELLE | - |
| IKKb (IKBKINASE) | O14920 | Y199 | ELLEQQKYTVTVDYW | + |
| IMP-1 | Q9NZI8 | Y396 | SVTGAAPYSSFMQAP | |
| iNOS | P35228 | Y151 | IEFVNQYYGSFKEAK | |
| ITGB3 | P05106 | Y773 | DTANNPLYKEATSTF | |
| ITGB3 | P05106 | Y785 | STFTNITYRGT____ | |
| ITGB7 | P26010 | Y753 | YRLSVEIYDRREYSR | |
| ITGB7 | P26010 | Y758 | EIYDRREYSRFEKEQ | |
| JIP3 | Q9UPT6 | Y598 | YPSVNIHYKSPTTAG | |
| JIP3 | Q9UPT6 | Y645 | REQKREQYRQVREHV | |
| JIP3 | Q9UPT6 | Y692 | KNVPVPVYCRPLVEK | |
| KDR (VEGFR2) | P35968 | Y1175 | AQQDGKDYIVLPISE | + |
| KHSRP | Q92945 | Y317 | GFGDRNEYGSRIGGG | |
| KHSRP | Q92945 | Y583 | WAAYYSHYYQQPPGP | |
| KHSRP | Q92945 | Y688 | YYRQQAAYYGQTPVP | |
| Kit | P10721 | Y900 | EHAPAEMYDIMKTCW | ? |
| Kv1.3 | P22001 | Y135 | FSEEIRFYQLGEEAM | |
| Kv1.3 | P22001 | Y447 | EGEEQSQYMHVGSCQ | |
| Kv2.1 | Q14721 | Y128 | YWGIDEIYLESCCQA | |
| Kv7.3 | O43525 | Y104 | VKRNNAKYRRIQTLI | |
| Kv7.3 | O43525 | Y386 | IQAAWRYYATNPNRI | |
| LMR1 (AATK) | Q6ZMQ8 | Y73 | ENAEGDEYAADLAQG | |
| LMR1 (AATK) iso2 | Q6ZMQ8 | Y93 | AQNGPDVYVLPLTEV | |
| LRP1 | Q07954 | Y4473 | VEIGNPTYKMYEGGE | |
| LRP1 | Q07954 | Y4507 | TNFTNPVYATLYMGG | |
| MPZL1 | O95297 | Y241 | SHQGPVIYAQLDHSG | |
| MPZL1 | O95297 | Y263 | NKSESVVYADIRKN_ | |
| MTSS1 | O43312 | Y393 | HLPDYAHYYTIGPGM | |
| MTSS1 | O43312 | Y394 | LPDYAHYYTIGPGMF | |
| MUC1 | P15941 | Y1229 | SSTDRSPYEKVSAGN | ? |
| MYLK1 (smMLCK) | Q15746 | Y464 | QEGSIEVYEDAGSHY | + |
| MYLK1 (smMLCK) | Q15746 | Y471 | YEDAGSHYLCLLKAR | + |
| N-CAM L1 | P32004 | Y1151 | KRSKGGKYSVKDKED | |
| N-CAM L1 | P32004 | Y1176 | KDETFGEYRSLESDN | |
| nAChRA7 | P36544 | Y386 | ASNGNLLYIGFRGLD | |
| nAChRA7 | P36544 | Y442 | KILEEVRYIANRFRC | |
| Nectin 2 | Q92692 | Y513 | LDKINPIYDALSYSS | |
| NMDAR1 (Glutamate) | Q05586 | Y837 | LIFIEIAYKRHKDAR | ? |
| Occludin | Q16625 | Y398 | KRTEQDHYETDYTTG | |
| Occludin | Q16625 | Y402 | QDHYETDYTTGGESC | |
| P130Cas (BCAR1) | P56945 | Y128 | SKAQQGLYQVPGPSP | |
| P130Cas (BCAR1) | P56945 | Y165 | PSPATDLYQVPPGPG | |
| P130Cas (BCAR1) | P56945 | Y234 | AQPEQDEYDIPRHLL | |
| P130Cas (BCAR1) | P56945 | Y249 | APGPQDIYDVPPVRG | |
| P130Cas (BCAR1) | P56945 | Y306 | PSNHHAVYDVPPSVS | |
| P130Cas (BCAR1) | P56945 | Y410 | GVVDSGVYAVPPPAE | |
| P130Cas (BCAR1) | P56945 | Y664 | EGGWMEDYDYVHLQG | |
| p27Kip1 | P46527 | Y74 | HKPLEGKYEWQEVEK | ? |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
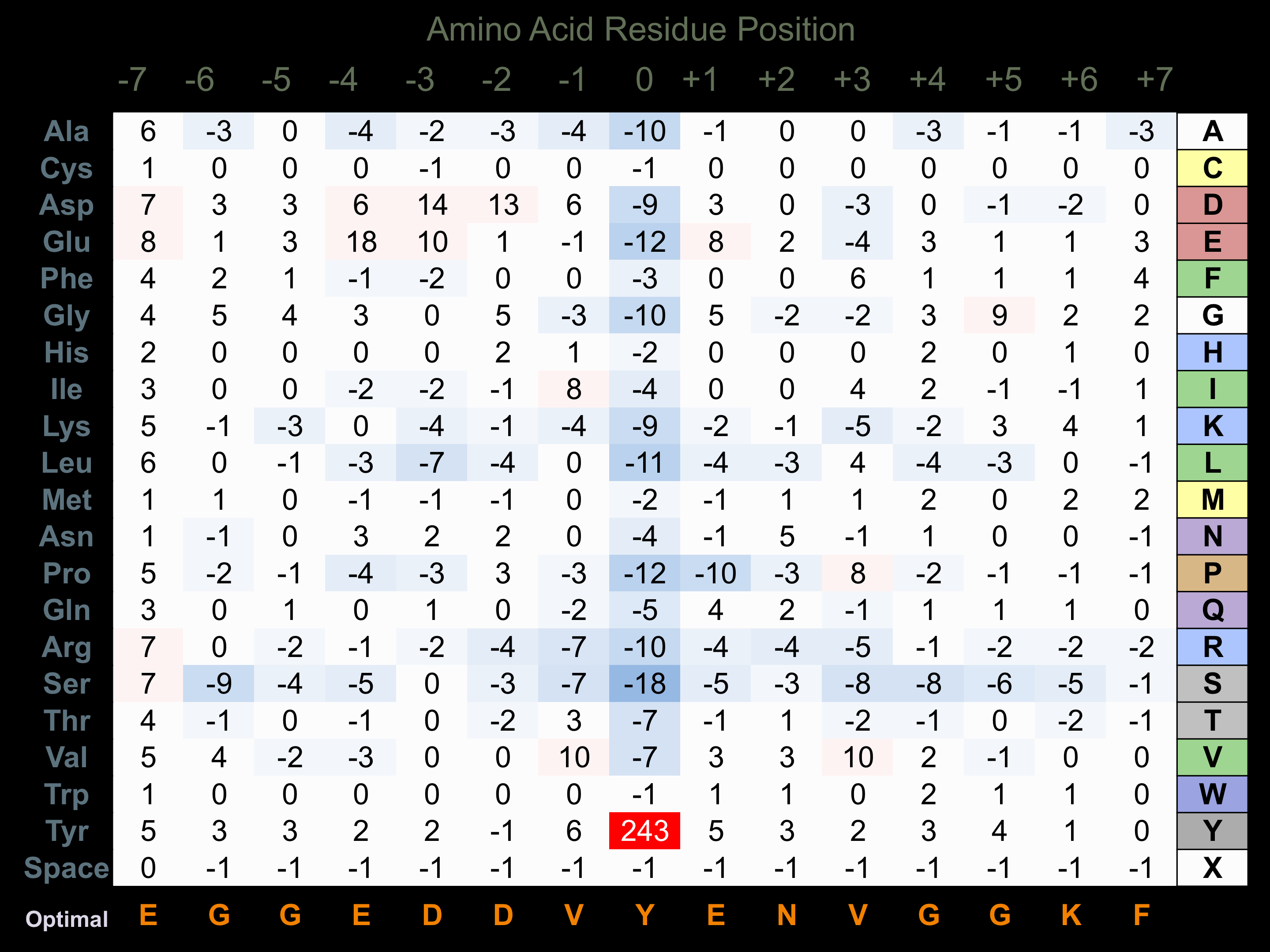
Matrix Type:
Experimentally derived from alignment of 522 known protein substrate phosphosites and 23 peptides phosphorylated by recombinant Src in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, immune disorders
Specific Diseases (Non-cancerous):
Influenza (FLU); Pneumococcal meningitis
Comments:
Pneumococcal Meningitis is a rare infectious disorder caused by streptococcus pneumonia, and an important gene related to this disorder is Src.
Specific Cancer Types:
Sarcomas; Colon cancer; Breast cancer; Agammaglobulinemias (IGHM); Cherubism (CRBM); Estrogen-receptor negative breast cancer; Differentiating neuroblastomas; Colon cancer, advanced, somatic
Comments:
SRC is a known oncoprotein (OP). Although mutations that increase the catalytic activity of SRC are well known, such as loss of the inhibitory tyrosine phosphorylation site at the C-terminus of the kinase, it is not subject to higher rates of mutations in human tumours than most proteins. The active form of the protein kinase normally acts to promote tumour cell proliferation. Agammaglobulinemia (IGHM) is considered a rare cancer immune disease which can be characterized by liver enlargement, monoclonal immunoglobulin, and infiltration of the bone marrow. Cherubism (CRBM) is a rare cancer characterized by malformation of bones in the lower portion of the face. CRBM results in bone being replaced by cyst-like growths resulting in enlarged features. CRBM can affect bone, B cells, and testes.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Cervical cancer stage 2A (%CFC= -67, p<0.083). The COSMIC website notes an up-regulated expression score for SRC in diverse human cancers of 734, which is 1.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 43 for this protein kinase in human cancers was 0.7-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. Src phosphotransferase activity can be abrogated with a K298M mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 27329 diverse cancer specimens. This rate is only -12 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.32 % in 809 skin cancers tested; 0.27 % in 2249 large intestine cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: Q531* (11).
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.