Nomenclature
Short Name:
FLT1
Full Name:
Vascular endothelial growth factor receptor 1
Alias:
- EC 2.7.1.112
- FRT
- Tyrosine-protein kinase FRT
- Tyrosine-protein kinase receptor FLT
- VGFR1
- EC 2.7.10.1
- FLT
- Flt-1
- Fms-like tyrosine kinase 1
- Fms-related tyrosine kinase 1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
VEGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
150769
# Amino Acids:
1338
# mRNA Isoforms:
8
mRNA Isoforms:
150,769 Da (1338 AA; P17948); 82,124 Da (733 AA; P17948-3); 77,474 Da (687 AA; P17948-2); 62,954 Da (556 AA; P17948-5); 60,917 Da (541 AA; P17948-4); 52,613 Da (463 AA; P17948-6); 41,175 Da (361 AA; P17948-8); 39,148 Da (343 AA; P17948-7)
4D Structure:
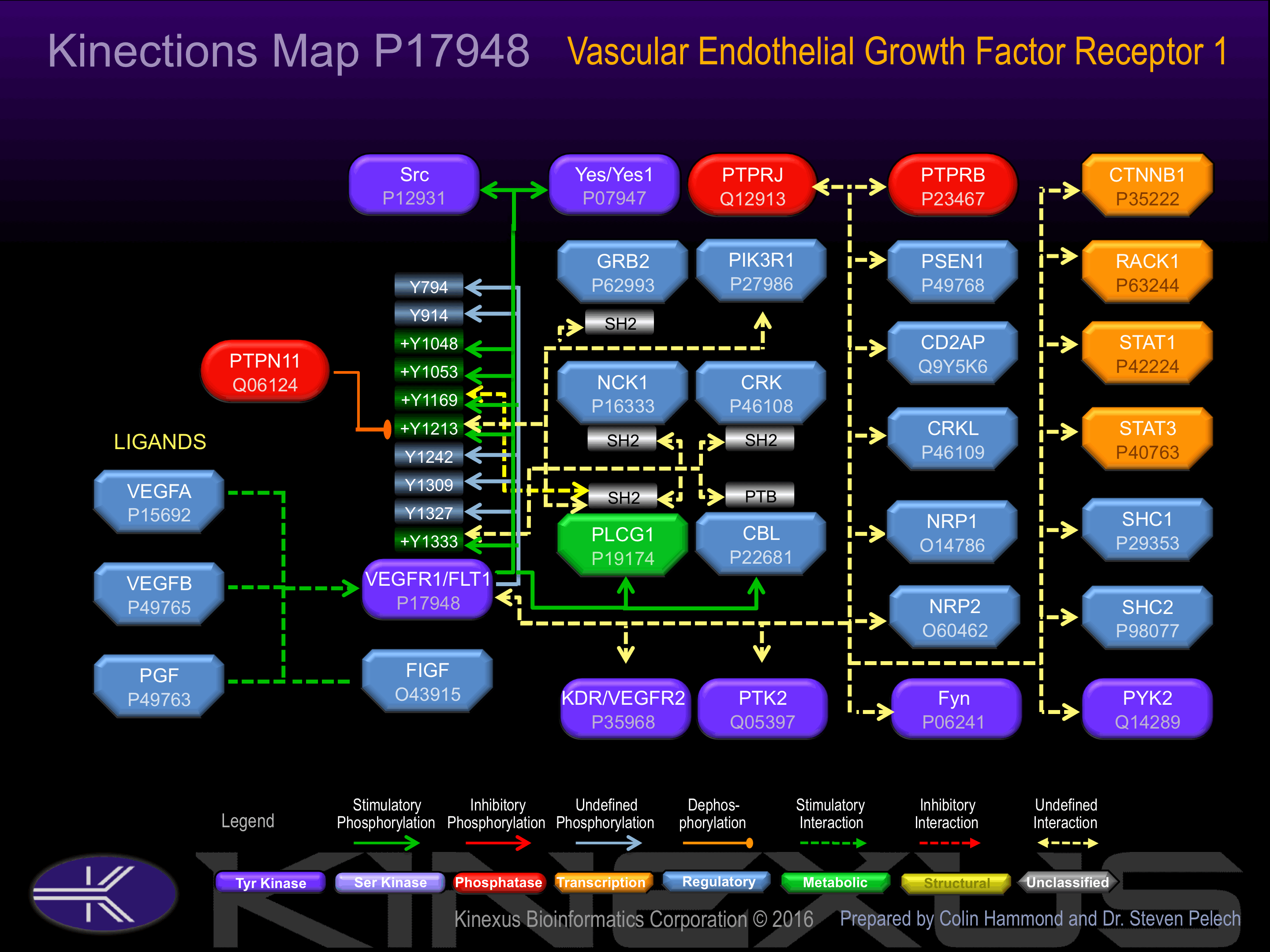
Interacts in vitro with various phosphotyrosine-binding proteins, including PLC-gammas, PTPN11, GRB2, CRK and NCK1.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
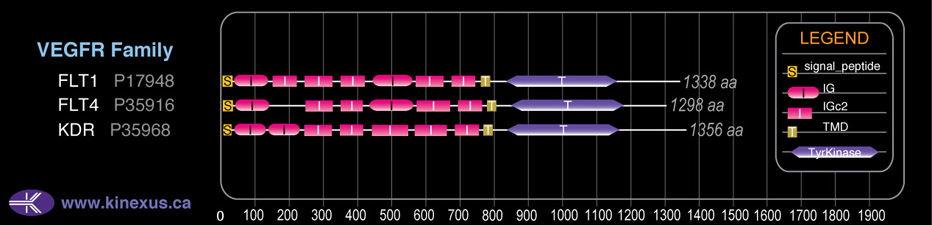
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
K306, K831, K932, K1153.
N-GlcNAcylated:
N100, N164, N196, N251, N323, N402, N417, N474, N547, N597, N620, N625, N666.
Serine phosphorylated:
S267, S631, S739, S742, S743, S1197, S1199, S1205, S1207.
Threonine phosphorylated:
T218, T222, T255, T265, T404, T588, T599, T618, T633.
Tyrosine phosphorylated:
Y383, Y388, Y745, Y794, Y815, Y911, Y920, Y1048+, Y1053+, Y1169+, Y1213+, Y1242, Y1327, Y1333+.
Ubiquitinated:
K410.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1227
63
1269
 4
4
43
31
37
 26
26
321
32
1305
 28
28
340
256
1409
 64
64
791
59
559
 6
6
69
158
164
 27
27
328
93
540
 48
48
592
108
1476
 26
26
313
24
238
 14
14
168
277
251
 4
4
49
80
55
 36
36
440
400
543
 5
5
59
65
79
 3
3
32
26
20
 10
10
117
74
403
 3
3
35
42
27
 13
13
154
798
1680
 26
26
319
51
1833
 7
7
91
231
249
 52
52
637
246
636
 15
15
184
71
957
 10
10
122
73
280
 12
12
153
51
323
 5
5
66
53
195
 17
17
205
71
1426
 64
64
788
170
1124
 13
13
159
68
843
 13
13
165
53
847
 19
19
229
52
524
 7
7
85
84
78
 22
22
274
30
338
 34
34
412
60
397
 4
4
44
115
165
 58
58
711
166
660
 13
13
163
87
197
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.3
99.3
99.9
99 88.3
88.3
89.3
98 -
-
-
84 -
-
-
95 89.9
89.9
95.4
90 -
-
-
- 81.7
81.7
90.4
82 82.4
82.4
90.7
82 -
-
-
- 71.9
71.9
83.9
- 69
69
80.4
71 25.6
25.6
41.7
49 44.2
44.2
61.4
54 28.7
28.7
45.1
- -
-
-
- -
-
-
- -
-
-
23 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by binding vascular endothelial growth factor (VEGF), which induces dimerization and autophosphorylation. Autophosphorylation of Tyr-1169 induces interaction with PLCg1. Autophosphorylation of Tyr-1213 increases phosphotransferase activity and induces interaction with Grb2, PLCg1 and SHP2. Autophosphorylation of Tyr-1333 induces interaction with Cbl, Crk, Nck1 and PLCg1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| FLT1 (VEGFR1) | P17948 | Y1053 | DIYKNPDYVRKGDTR | |
| FLT1 (VEGFR1) | P17948 | Y1169 | VQQDGKDYIPINAIL | + |
| FLT1 (VEGFR1) | P17948 | Y1213 | GSSDDVRYVNAFKFM | + |
| FLT1 (VEGFR1) | P17948 | Y1242 | ATSMFDDYQGDSSTL | ? |
| FLT1 (VEGFR1) | P17948 | Y1309 | EGKRRFTYDHAELER | |
| FLT1 (VEGFR1) | P17948 | Y1327 | CCSPPPDYNSVVLYS | |
| FLT1 (VEGFR1) | P17948 | Y1333 | DYNSVVLYSTPPI__ | + |
| FLT1 (VEGFR1) | P17948 | Y914 | VIVEYCKYGNLSNYL |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
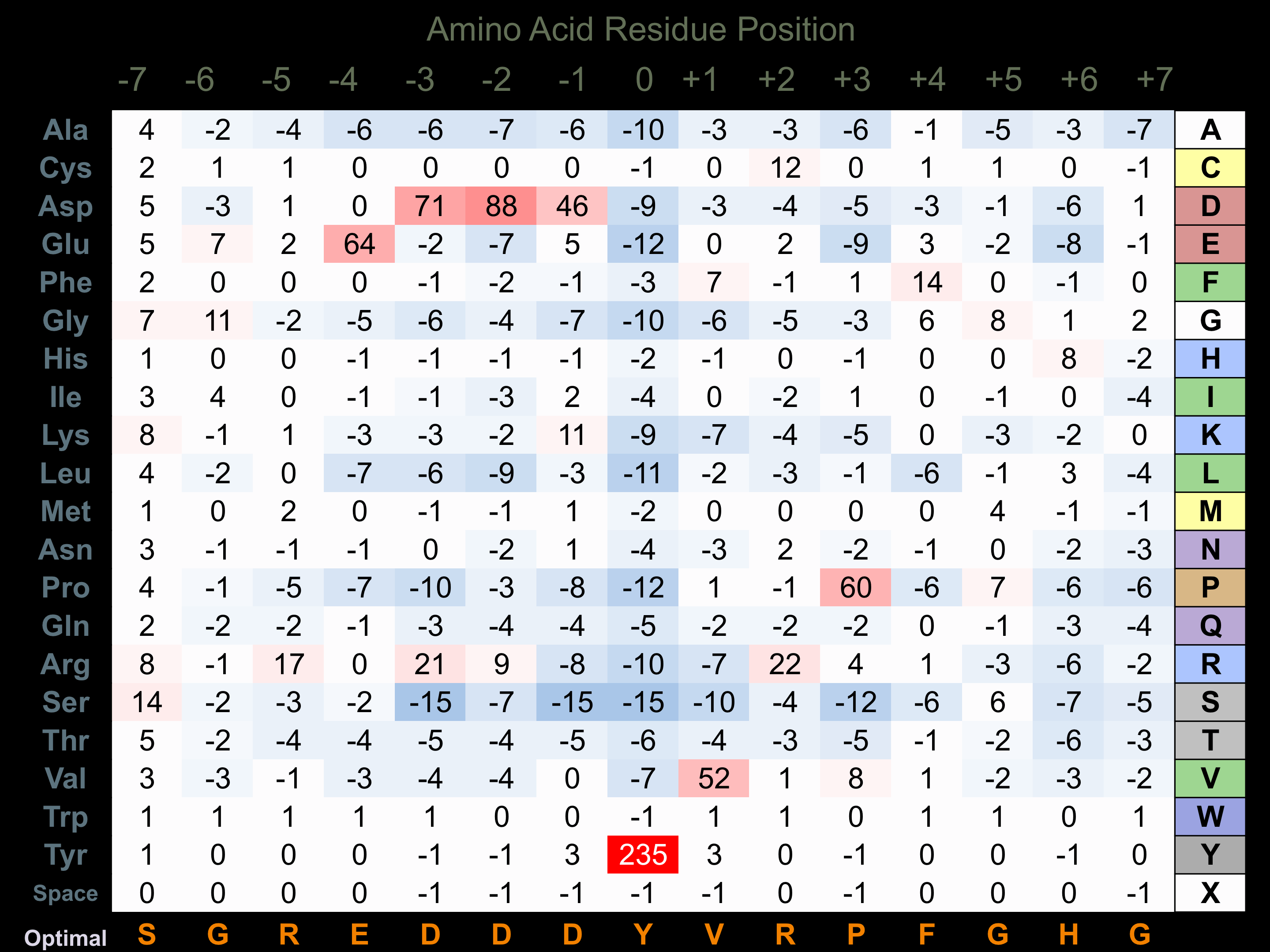
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, reproductive disorders
Specific Diseases (Non-cancerous):
Pre-eclampsia; Eclampsia; Placental insufficiency; Acute mountain sickness; Lipodermatosclerosis
Comments:
Pre-eclampsia is a pregancy complication characterized by the maternal development of high blood pressure (hypertension) and high levels of protein in the urine. This complication usually occurs in the last few months of the pregnancy and commonly results in the premature delivery of the infant. Placental insufficiency is a pregnancy complication characterized by insufficient blood flow to the placenta, which can results in fetal distress and a range of other pregnancy complications, including fetal death. Mutations in FLT1 are associated with these disorders. In women with pre-eclampsia, increased expression of FLT1 correlated with decreased levels of circulating VEGF and PGF, which is predicted to cause defects in the endothelial cells of the blood vessels. In animal studies, administration of the FLT1 protein to pregnant rats resulted in the development of hypertension, proteinuria (protein in the urine), and glomerula endothliosis, which closely resembled the pre-eclampsia phenotype in humans. Therefore, increased activity and/or expression of the FLT1 protein has been implicated in the pathogenesis of pre-eclampsia and placental insufficiently.
Specific Cancer Types:
Breast cancer; Meningioma; Astrocytoma; Epithelioid hemangioendothelioma; Cancer-associated retinopathy; Microcystic meningioma
Comments:
FLT1 appears to be an oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. Gain-of-function mutations in the FLT1 gene are associated with cancer development. FLT1 expression has been shown to contribute to cancer cell survival, proliferation, migration, tissue invasion, tumour angiogenesis, and metastasis. In addition, FLT1 may promote cancer pathogenesis by enhancing inflammatory responses and recruiting tumor-infiltrating macrophages.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Clear cell renal cell carcinomas (cRCC) (%CFC= +288, p<0.0006); Clear cell renal cell carcinomas (cRCC) (%CFC= +130, p<0.075); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +311, p<0.009); Malignant pleural mesotheliomas (MPM) tumours (%CFC= -51, p<0.02); Ovary adenocarcinomas (%CFC= +139, p<0.0001); Skin melanomas (%CFC= -46, p<0.1); and Skin melanomas - malignant (%CFC= -52, p<0.045). The COSMIC website notes an up-regulated expression score for FLT1 in diverse human cancers of 347, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 25596 diverse cancer specimens. This rate is a modest 1.54-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.7 % in 856 skin cancers tested; 0.57 % in 1128 large intestine cancers tested; 0.34 % in 590 stomach cancers tested; 0.21 % in 1949 lung cancers tested; 0.19 % in 602 endometrium cancers tested; 0.12 % in 605 oesophagus cancers tested; 0.07 % in 1270 liver cancers tested; 0.07 % in 1226 kidney cancers tested; 0.05 % in 1963 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R781Q (9).
Comments:
Only 7 deletion, 2 insertions and no complex mutations are noted on the COSMIC website.