Nomenclature
Short Name:
FGFR2
Full Name:
Fibroblast growth factor receptor 2
Alias:
- BEK
- Crouzon syndrome
- EC 2.7.10.1
- ECT1
- K-SAM; TK25
- BFR1
- BFR-1
- CD332
- CEK3
- CFD1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
FGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
92,025
# Amino Acids:
821
# mRNA Isoforms:
23
mRNA Isoforms:
92,733 Da (830 AA; P21802-11); 92,153 Da (822 AA; P21802-16); 92,118 Da (822 AA; P21802-3); 92,025 Da (821 AA; P21802); 91,918 Da (820 AA; P21802-18); 91,825 Da (819 AA; P21802-5); 91,641 Da (819 AA; P21802-10); 91,620 Da (817 AA; P21802-7); 91,566 Da (819 AA; P21802-9); 88,181 Da (785 AA; P21802-6); 86,407 Da (771 AA; P21802-12); 86,223 Da (769 AA; P21802-17); 86,130 Da (768 AA; P21802-2); 86,110 Da (768 AA; P21802-13); 85,929 Da (766 AA; P21802-8); 79,833 Da (709 AA; P21802-23); 79,300 Da (707 AA; P21802-21); 79,212 Da (705 AA; P21802-15); 79,197 Da (704 AA; P21802-20); 76,705 Da (682 AA; P21802-4); 76,423 Da (680 AA; P21802-22); 40,614 Da (366 AA; P21802-19); 28,299 Da (254 AA; P21802-14)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K539.
N-GlcNAcylated:
N83, N123, N228, N241, N265, N297, N318, N331.
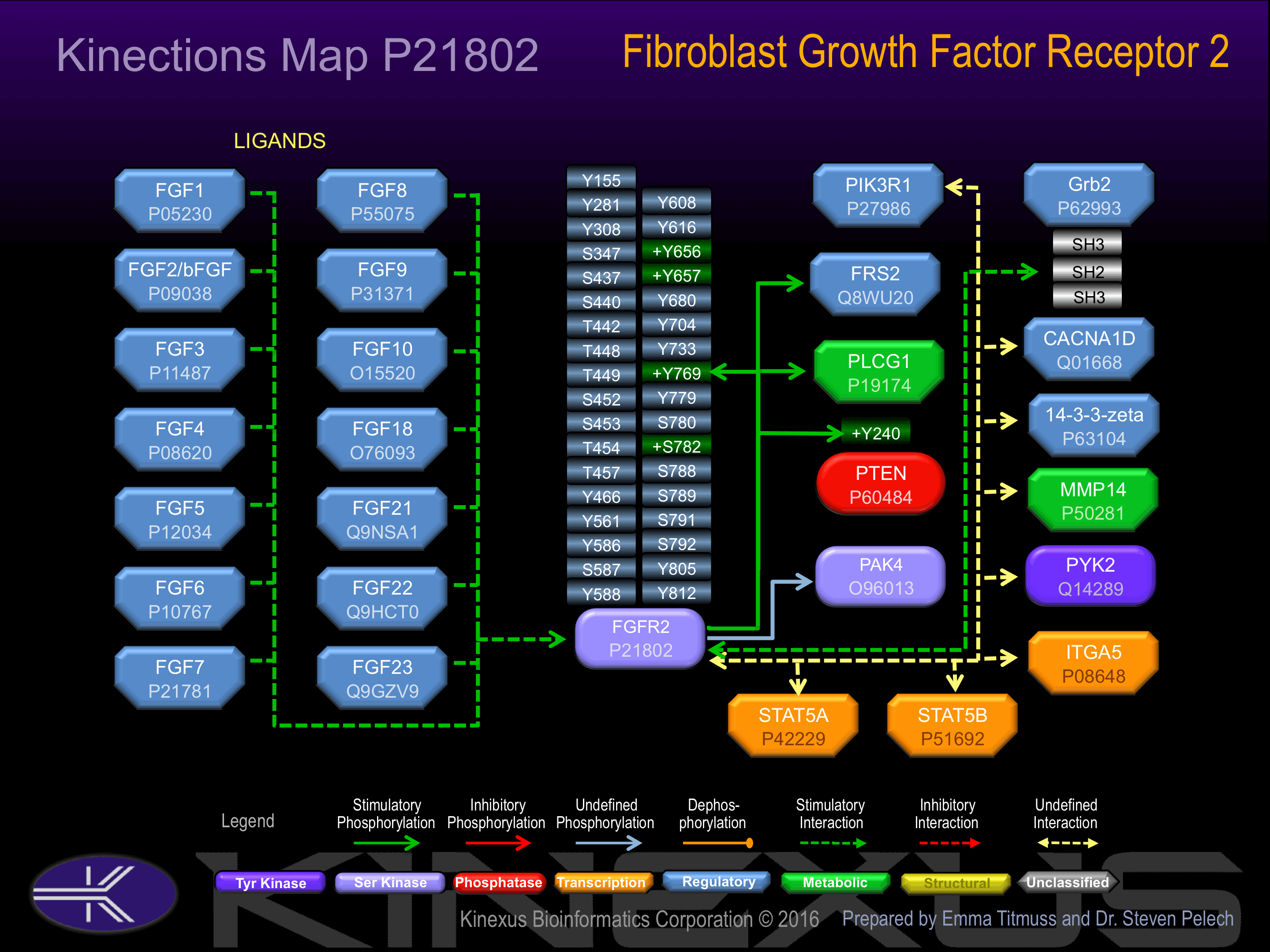
Serine phosphorylated:
S347, S437, S440, S452, S453, S533, S587, S780, S782, S788, S789, S791, S792.
Threonine phosphorylated:
T442, T448, T449, T454, T457.
Tyrosine phosphorylated:
Y466, Y586, Y588, Y608, Y616, Y656+, Y657+, Y733, Y769+, Y805, Y812.
Ubiquitinated:
K751.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
820
142
1194
 8
8
62
67
93
 15
15
127
58
146
 40
40
326
484
476
 79
79
646
122
549
 31
31
251
404
1209
 28
28
233
170
447
 75
75
612
180
1162
 52
52
427
76
381
 10
10
85
462
125
 12
12
96
146
144
 77
77
635
795
610
 3
3
23
169
43
 5
5
39
50
59
 12
12
98
115
145
 7
7
54
85
90
 9
9
71
679
83
 26
26
216
94
259
 3
3
26
466
53
 64
64
522
592
613
 64
64
526
109
692
 4
4
30
123
55
 10
10
86
77
81
 6
6
50
94
60
 7
7
60
107
75
 78
78
637
294
626
 7
7
60
174
104
 24
24
199
95
251
 25
25
207
95
247
 12
12
96
168
107
 1
1
8
60
9
 92
92
751
166
1316
 4
4
33
216
314
 89
89
726
317
634
 13
13
105
156
94
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.6
99.6
99.6
95 88.4
88.4
90.9
- -
-
-
96 -
-
-
97 -
-
-
99 -
-
-
- 97
97
98.2
95 71.8
71.8
83.8
94.5 -
-
-
- -
-
-
- 92.8
92.8
96.3
90.5 77.6
77.6
87.2
76 77.5
77.5
86.6
77 -
-
-
- 29.1
29.1
45.1
42 -
-
-
- 31.6
31.6
46.5
- 34.5
34.5
51.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | FGF4 - P08620 |
| 2 | FGF7 - P21781 |
| 3 | KGFLP1 - Q2TVT4 |
| 4 | FGF6 - P10767 |
| 5 | FGF8 - P55075 |
| 6 | KGFLP2 - Q2TVT3 |
| 7 | FGF10 - O15520 |
| 8 | FGF2 - P09038 |
| 9 | FGF23 - Q9GZV9 |
| 10 | FGF5 - P12034 |
| 11 | CACNA1D - Q01668 |
| 12 | ITGA5 - P08648 |
| 13 | FGF18 - O76093 |
| 14 | EPHA4 - P54764 |
| 15 | FRS2 - Q8WU20 |
Regulation
Activation:
Activated by binding of fibroblast growth factor (FGF), which induces dimerization and autophosphorylation. Phosphorylation of Tyr-769 increases phosphotransferase activity and induces interaction with FRS2 and PLCG1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
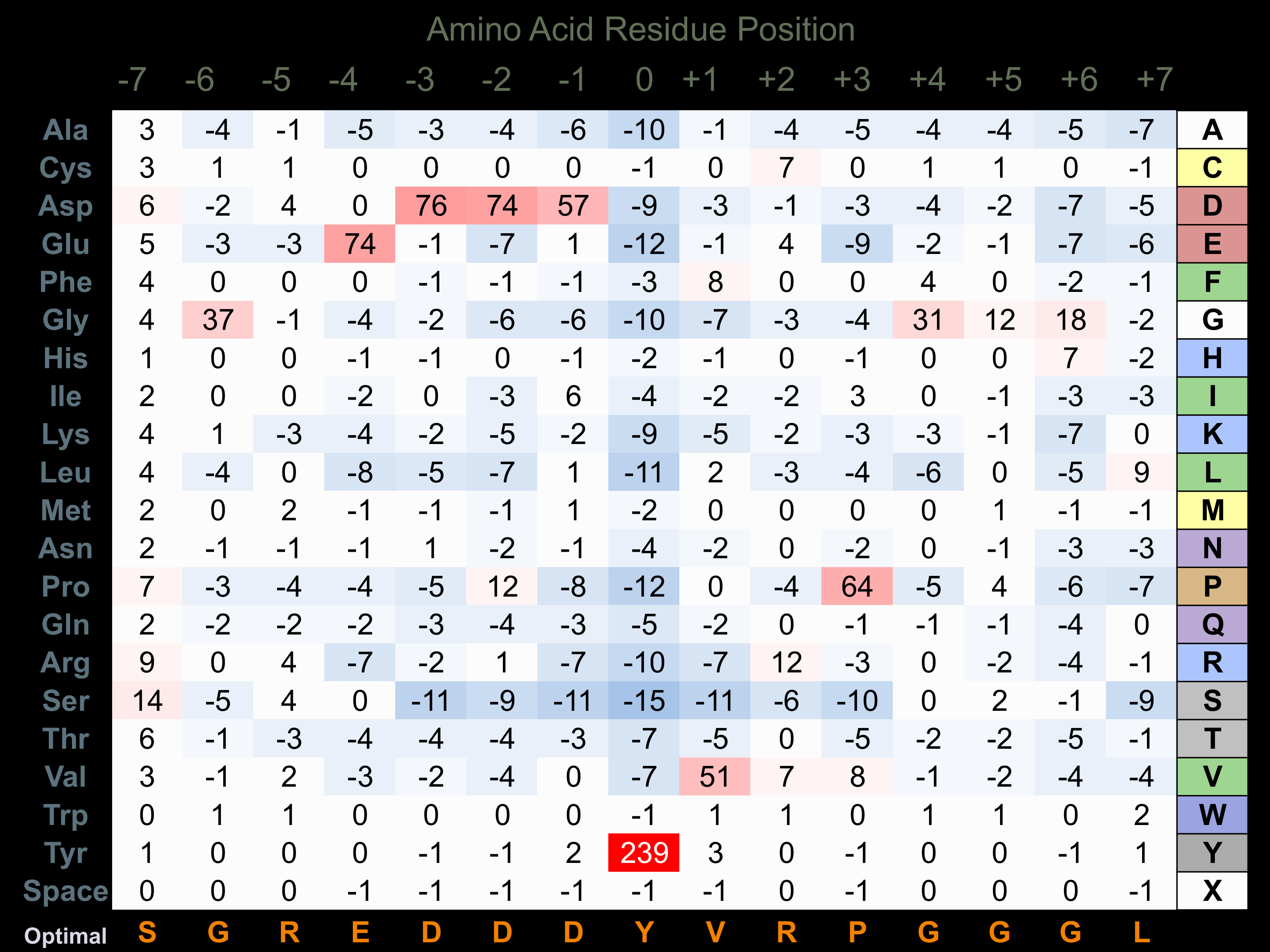
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, bone disorders, infectious disorders, eye disorders, and neuronal disorders
Specific Diseases (Non-cancerous):
Bent Bone dysplasia syndrome; Infectious mononucleosis; Crouzon syndrome; Jackson-Weiss syndrome (JWS); Beare-Stevenson Cutis Gyrata syndrome; Saethre-Chotzen syndrome (CS); Ladd syndrome; Craniosynostosis; Acrocephalosyndactylia; Achondroplasia; Antley-Bixler syndrome without genital anomalies or disordered steroidogenesis; Synostosis; Antley-Bixler syndrome; Acanthosis nigricans; Hypochondroplasia; Thanatophoric dysplasia; Exophthalmos; Pfeiffer syndrome (PS) Type 1; Scaphocephaly, Maxillary Retrusion, and Mental Retardation; Crouzonodermoskeletal syndrome; Plagiocephaly; Isolated Anorectal malformation; Humeroradial synostosis; Muenke syndrome; Wells syndrome; Syringomyelia; Thanatophoric dysplasia Type 1; Gliomatosis cerebri; Osteoglophonic dysplasia; Acanthoma; Radioulnar Synostosis; Cytochrome P450 oxidoreductase deficiency; Clear cell acanthoma; FGFR-related craniosynostosis syndromes; Pfeiffer syndrome Type 1, 2 and 3; Saethre-Chotzen syndrome, FGFR2-related; Craniosynostosis, Nonspecific; FGFR2-related isolated coronal synostosis; FGFR2-related lacrimo-auriculo-dento-digital syndrome; Scaphocephaly and Axenfeld-Rieger anomaly
Comments:
Mutations of the FGFR2 gene has been found to be associated with many developmental diseases in abnormal growth and potential mental retardation, such as Crouzon syndrome (CS), Jackson-Weiss syndrome (JWS), Apert syndrome (APRS), Pfeiffer syndrome (PS), and Beare-Stevenson cutis gyrata syndrome (BSTVS).
Specific Cancer Types:
Breast cancer; Colorectal cancer (CRC); Breast cancer susceptibility; Gastric cancer, somatic
Comments:
FGFR2 is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. SNP in intron 2 of FGFR2 has been shown to be significantly associated with breast cancer. An activating mutation of FGFR2 gene in gastric cancer has been found.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -51, p<0.029); Cervical cancer (%CFC= +282, p<0.001); Cervical cancer stage 2A (%CFC= -47, p<0.001); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -50, p<0.047); Lung adenocarcinomas (%CFC= -55, p<0.0001);Oral squamous cell carcinomas (OSCC) (%CFC= +165, p<0.003); Ovary adenocarcinomas (%CFC= +54, p<0.077); Skin melanomas (%CFC= -72, p<0.073); Skin melanomas - malignant (%CFC= -80, p<0.003); and Uterine leiomyomas (%CFC= -63, p<0.002). The COSMIC website notes an up-regulated expression score for FGFR2 in diverse human cancers of 370, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.17 % in 31153 diverse cancer specimens. This rate is 2.3-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.98 % in 1436 endometrium cancers tested; 0.71 % in 1367 large intestine cancers tested; 0.58 % in 1527 skin cancers tested; 0.26 % in 886 stomach cancers tested; 0.15 % in 2356 lung cancers tested; 0.13 % in 1270 liver cancers tested; 0.1 % in 1876 breast cancers tested; 0.06 % in 2123 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S252W (49); N549K (30); C382R (12); Y375C (10).
Comments:
Only 5 deletions, 2 insertions and 1 complex mutation noted on the COSMIC website.