Nomenclature
Short Name:
TYK2
Full Name:
Non-receptor tyrosine-protein kinase TYK2
Alias:
- EC 2.7.10.2
- JTK1
- Kinase Tyk2
- Tyrosine kinase 2
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
JakA
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
133,650
# Amino Acids:
1187
# mRNA Isoforms:
1
mRNA Isoforms:
133,650 Da (1187 AA; P29597)
4D Structure:
Interacts with JAKMIP1
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
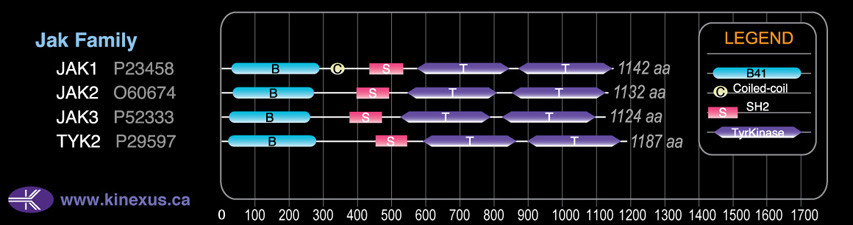
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S339, S340, S491, S499, S525, S577, S720, S723, S884.
Threonine phosphorylated:
T67.
Tyrosine phosphorylated:
Y292, Y423, Y433, Y604, Y827, Y851, Y1054+, Y1055+, Y1145.
Ubiquitinated:
K756.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 31
31
927
16
1152
 4
4
121
10
42
 3
3
76
1
0
 28
28
822
53
980
 27
27
810
14
770
 68
68
2029
45
4495
 14
14
410
19
615
 71
71
2115
28
3484
 22
22
661
10
560
 6
6
180
43
117
 5
5
150
12
50
 23
23
680
105
723
 6
6
167
12
47
 2
2
74
9
48
 3
3
102
11
56
 4
4
133
8
21
 13
13
396
102
2241
 3
3
83
8
33
 4
4
134
46
96
 23
23
700
56
717
 4
4
127
10
56
 9
9
269
12
149
 8
8
239
10
138
 1
1
30
8
13
 8
8
242
10
169
 100
100
2989
35
3427
 6
6
184
15
51
 3
3
94
8
30
 4
4
107
8
18
 4
4
119
14
84
 47
47
1418
18
874
 23
23
678
21
731
 9
9
279
57
483
 30
30
910
31
762
 4
4
130
22
111
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 21.1
21.1
36.9
99 82.1
82.1
85.8
95 -
-
-
83 -
-
-
87 52.7
52.7
57.1
83 -
-
-
- 79.5
79.5
86.7
80.5 38.4
38.4
55.6
81 -
-
-
- -
-
-
- 38.2
38.2
55.3
67 59
59
74.1
62 45.7
45.7
62.8
53 -
-
-
- 22.9
22.9
38.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PTPN6 - P29350 |
| 2 | PLAUR - Q03405 |
| 3 | STAT1 - P42224 |
| 4 | PTPRC - P08575 |
| 5 | FYN - P06241 |
| 6 | IL13RA1 - P78552 |
| 7 | CBL - P22681 |
| 8 | PTPN1 - P18031 |
| 9 | CRKL - P46109 |
| 10 | IRS1 - P35568 |
| 11 | PIK3R1 - P27986 |
| 12 | IRS2 - Q9Y4H2 |
| 13 | STAT2 - P52630 |
| 14 | PTAFR - P25105 |
| 15 | STAM2 - O75886 |
Regulation
Activation:
Phosphorylation of Tyr-1054 and Tyr-1055 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
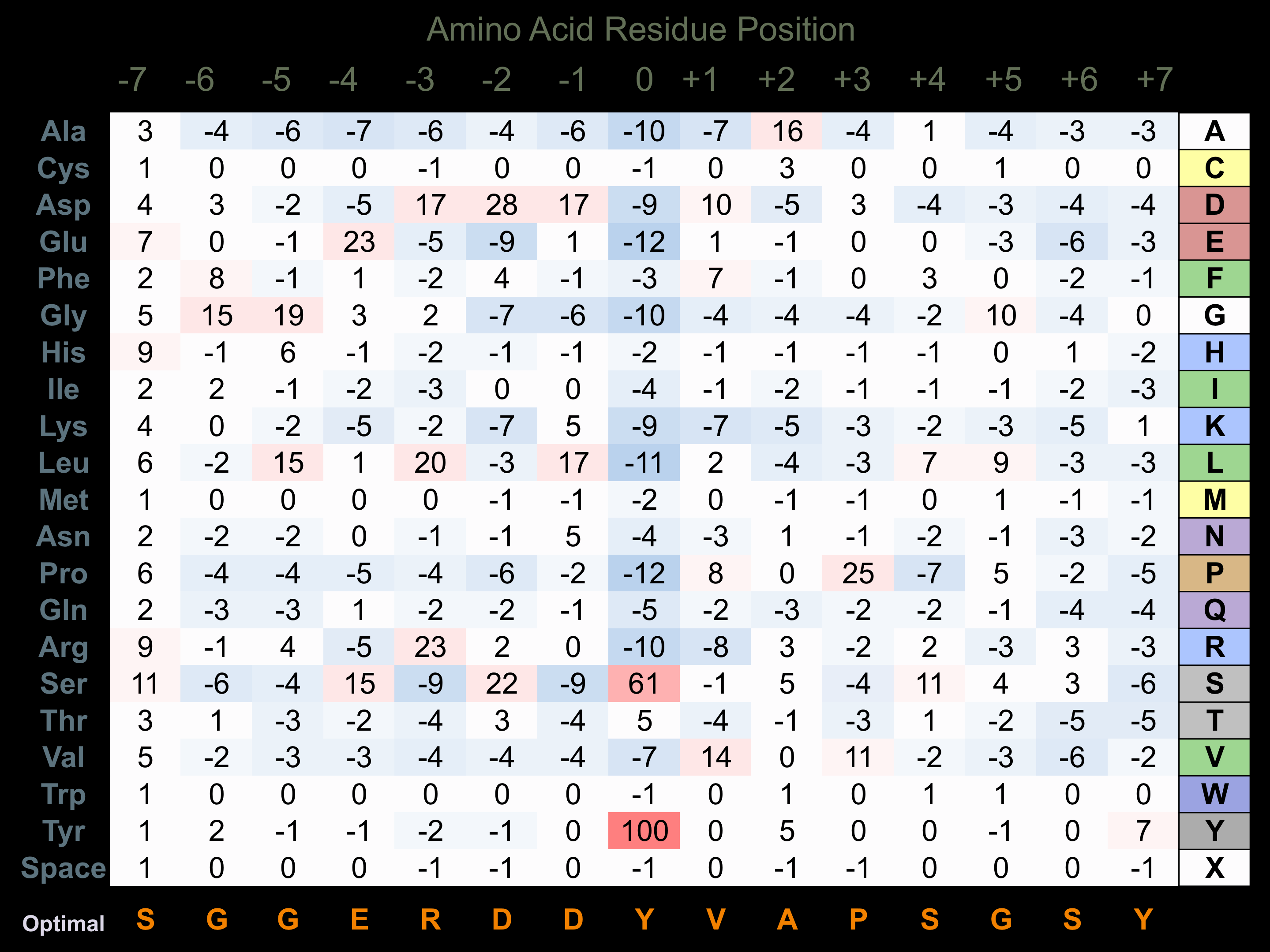
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
2
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, immune, respiratory, bone, and skin disorders
Specific Diseases (Non-cancerous):
Protein-tyrosine kinase 2 deficiency (TYK2 deficiency); Hyper Ige syndrome (HIES); Venezuelan equine encephalitis; Subacute cutaneous lupus erythematosus (SCLE); Polyarticular onset juvenile Idiopathic arthritis; Oligoarticular juvenile arthritis; Familial atypical mycobacteriosis, Tyk2-related
Comments:
Protein-tyrosine kinase 2 deficiency (TYK2 deficiency) is a disorder consisting of a defective immune system and is characterized by pneumonia, skin abscesses, and high levels of IgE in serum. Hyper Ige Syndrome (HIES) is a rare immunodeficiency disorder where serum IgE may be elevated, and the skin may be inflamed. One form of HIES, AD-HIES, is characterized by connective tissue issues, skeletal deformations, distinct facial structure, and dental abnormalities. The AR-HIES form of the disease is characterized by extreme hypereosinophilia, viral infection susceptibility, an abnormal central nervous system, and T-cell defects. Venezuelan Equine Encephalitis is a rare infectious disorder which is characterized by inflammation of the brain in humans or horses. Subacute Cutaneous Lupus Erythematosus (SCLE) is a rare immune and skin disorder that results in scaled lesions on skin. SCLE can affect skin, T cells, and lung tissues. Polyarticular Onset Juvenile Idiopathic Arthritis is a rare respiratory and bone disorder which has been related to Juvenile Rheumatoid Arthritis and Rheumatoid Arthritis. Some symptoms of Polyarticular Onset Juvenile Idiopathic Arthritis include biological inflammatory syndrome, upregulated erythrocyte sedimentation rate, and autoimmunity. Oligoarticular Juvenile Arthritis is characterized by joint pain, hydrarthrosis, and autoimmunity. Oligoarticular Juvenile Arthritis can affect the eye, bone, and T cells.
Specific Cancer Types:
Polycythemia vera (PV); Gallbladder adenocarcinomas
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +98, p<0.025); Bladder carcinomas (%CFC= +59, p<0.002); Brain glioblastomas (%CFC= -97, p<0.0001); Brain oligodendrogliomas (%CFC= -90, p<0.0001); Breast epithelial cell carcinomas (%CFC= +60, p<0.003); Cervical cancer (%CFC= -59, p<0.0001); Classical Hodgkin lymphomas (%CFC= +66, p<0.0005); Large B-cell lymphomas (%CFC= +136, p<0.0004); Ovary adenocarcinomas (%CFC= +65, p<0.032); Skin fibrosarcomas (%CFC= +65); and Skin melanomas - malignant (%CFC= +86, p<0.0001). The COSMIC website notes an up-regulated expression score for TYK2 in diverse human cancers of 544, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 151 for this protein kinase in human cancers was 2.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25688 diverse cancer specimens. This rate is very similar (+ 3% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.29 % in 805 skin cancers tested; 0.29 % in 1093 large intestine cancers tested; 0.22 % in 602 endometrium cancers tested; 0.19 % in 589 stomach cancers tested; 0.19 % in 500 urinary tract cancers tested; 0.14 % in 605 oesophagus cancers tested; 0.1 % in 1941 lung cancers tested; 0.09 % in 904 ovary cancers tested; 0.06 % in 1522 breast cancers tested; 0.05 % in 2228 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: N346T (4); G422S (4).
Comments:
Only 1 deletion, 3 insertions and 1 complex mutations are noted on the COSMIC website.

