Nomenclature
Short Name:
PDK1
Full Name:
3-phosphoinositide dependent protein kinase-1
Alias:
- 3-phosphoinositide dependent protein kinase-1
- MPDK1
- PDPK1
- PkB kinase
- PkB-like; PkB-like 2; PRO0461; Protein kinase B kinase
- EC 2.7.11.1
- HPDK1
- Kinase PDK1
- MGC20087
- MGC35290
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKB
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
# Amino Acids:
556
# mRNA Isoforms:
5
mRNA Isoforms:
63,152 Da (556 AA; O15530); 60,396 Da (530 AA; O15530-3); 58,035 Da (506 AA; O15530-2); 50,838 Da (454 AA; O15530-5); 48,201 Da (429 AA; O15530-4)
4D Structure:
Interacts with NPRL2
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
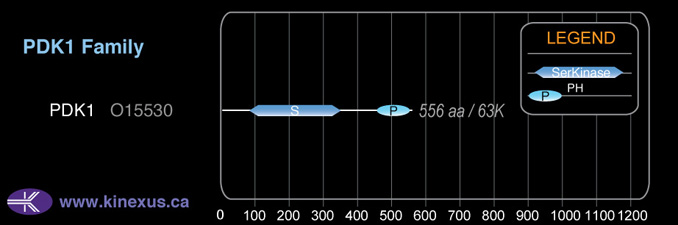
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K304 (N6)K534.
Serine phosphorylated:
S22, S25, S160+, S176, S231+, S234, S241+, S366, S393, S394, S396, S398, S410, S501, S529.
Threonine phosphorylated:
T33, T37, T54, T95, T148, T180, T245-, T255, T354, T513+.
Tyrosine phosphorylated:
Y9+, Y126, Y156, Y161, Y248, Y288, Y373+, Y376+, Y485, Y486.
Ubiquitinated:
K304, K441.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 58
58
1034
48
1062
 11
11
197
21
143
 9
9
161
10
167
 24
24
422
162
481
 49
49
867
46
729
 13
13
224
134
664
 15
15
276
53
451
 92
92
1633
61
3114
 27
27
483
24
420
 13
13
223
136
195
 8
8
144
39
126
 43
43
775
228
672
 4
4
77
43
92
 5
5
92
15
75
 7
7
128
23
125
 7
7
132
27
103
 9
9
168
213
250
 12
12
216
21
225
 9
9
161
145
151
 38
38
676
190
661
 10
10
183
27
160
 8
8
146
33
127
 14
14
241
13
234
 14
14
255
21
232
 9
9
169
27
140
 100
100
1784
97
3780
 5
5
98
46
102
 8
8
135
21
91
 8
8
144
21
110
 12
12
221
56
200
 29
29
524
30
618
 38
38
683
56
618
 10
10
170
101
314
 61
61
1097
104
860
 40
40
718
61
539
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
97 -
-
-
- -
-
-
96 -
-
-
96 93.3
93.3
95
95 -
-
-
- 94.8
94.8
96.2
95 95.3
95.3
96.4
96 -
-
-
- 52.1
52.1
54.6
- 28.6
28.6
44
90 28
28
43.8
86 27.5
27.5
43.3
81 -
-
-
- 32.5
32.5
44.1
51 54.1
54.1
66.1
- 32.5
32.5
52.2
43 58
58
70.3
- -
-
-
- -
-
-
- -
-
-
44 -
-
-
44 23.7
23.7
35.9
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | AKT1 - P31749 |
| 2 | RPS6KB1 - P23443 |
| 3 | AKT3 - Q9Y243 |
| 4 | TTC5 - Q8N0Z6 |
| 5 | PAK1 - Q13153 |
| 6 | ITGB3 - P05106 |
| 7 | SGK3 - Q96BR1 |
| 8 | YWHAH - Q04917 |
| 9 | PRKCB - P05771 |
| 10 | PRKCE - Q02156 |
| 11 | AKTIP - Q9H8T0 |
| 12 | IRS1 - P35568 |
| 13 | AKT2 - P31751 |
| 14 | PRKACA - P17612 |
| 15 | PXN - P49023 |
Regulation
Activation:
Activated by phosphorylaton at Tyr-9, Ser-160, Ser-241, Tyr-373, and Tyr-376.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| RET | P07949 | Y9 | ARTTSQLYDAVPIQS | + |
| SRC | P12931 | Y9 | ARTTSQLYDAVPIQS | + |
| PDK1 | O15530 | S25 | VVLCSCPSPSMVRTQ | |
| PDK1 | O15530 | S241 | SKQARANSFVGTAQY | + |
| SRC | P12931 | Y373 | SEDDEDCYGNYDNLL | + |
| SRC | P12931 | Y376 | DEDCYGNYDNLLSQF | + |
| PDK1 | O15530 | S393 | MQVSSSSSSHSLSAS | |
| PDK1 | O15530 | S396 | SSSSSSHSLSASDTG | ? |
| PDK1 | O15530 | S410 | GLPQRSGSNIEQYIH | |
| PKCt | Q04759 | S501 | LKGEIPWSQELRPEA | |
| PDK1 | O15530 | T513 | PEAKNFKTFFVHTPN | + |
| PKCt | Q04759 | S529 | TYYLMDPSGNAHKWC |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
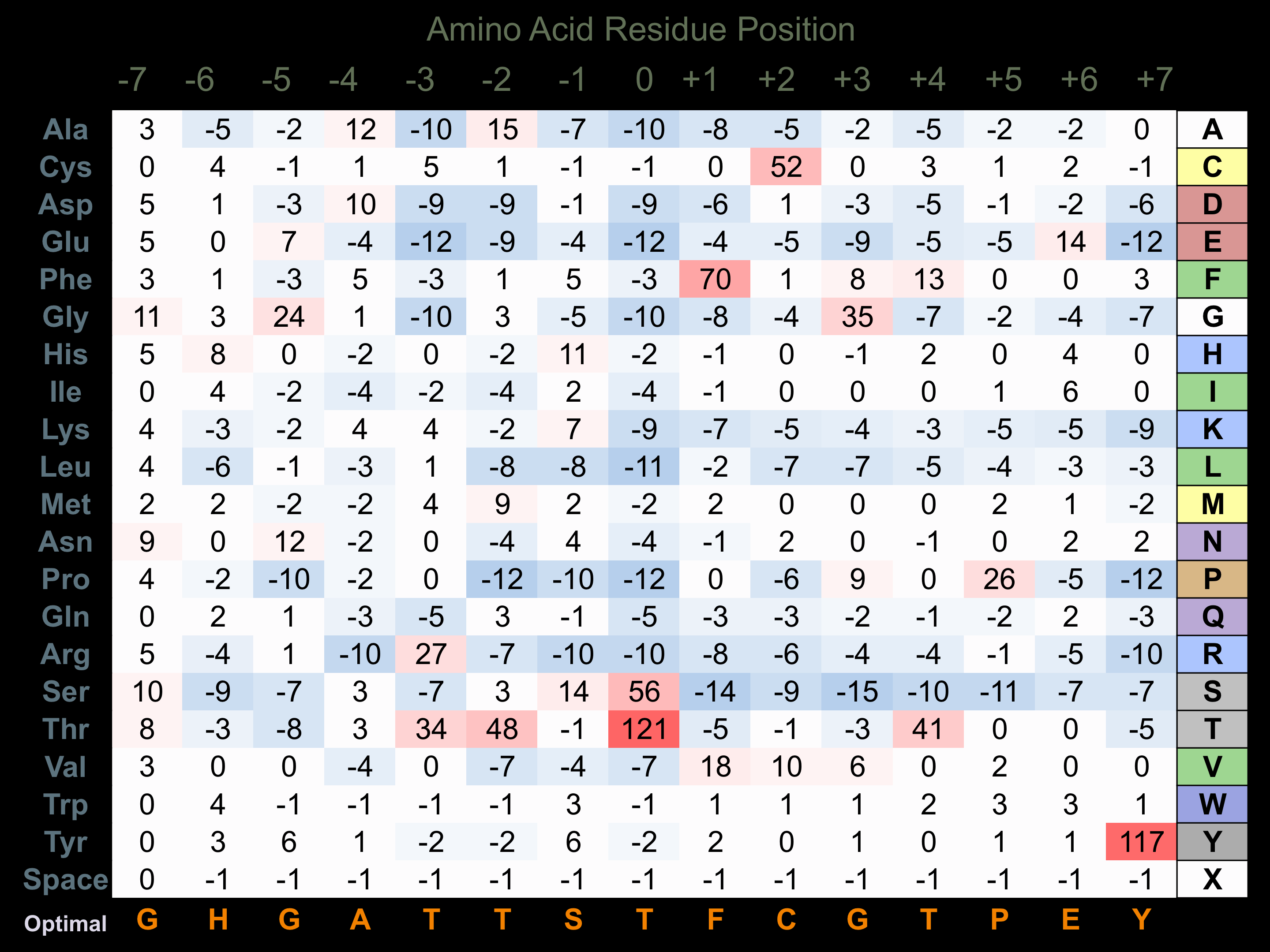
Matrix Type:
Experimentally derived from alignment of 62 known protein substrate phosphosites and 87 peptides phosphorylated by recombinant PDK1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Breast cancer
Comments:
PDK1 is important in mediating migration and metastasis of breast cancer cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +361, p<0.002); and Skin melanomas - malignant (%CFC= +85, p<0.001). The COSMIC website notes an up-regulated expression score for PDK1 in diverse human cancers of 439, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 166 for this protein kinase in human cancers was 2.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. Tyrosine phosphorylation induced by pervanadate in PDK1 can be partly reduced through a Y9F mutation in PDK1. Activation of PDK1 can be inhibited through a S241A mutation. PDK1 phosphotransferase activity can be reduced 3-fold with a A277V mutation. MELK-mediated phosphorylation can be inhibited through a T354A mutation. General phosphotransferase activity can also be reduced through either a Y373F, or a Y376F mutation. PDK1 phosphorylation of MAP3K5 can be ablated through the S394A and S398A mutations. Membrane localization via PDGF can be inhibited through a R474A mutation. Akt phosphorylation by PDK1 can be increased with a T513E mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.04 % in 25273 diverse cancer specimens. This rate is -46 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.17 % in 1270 large intestine cancers tested; 0.16 % in 895 skin cancers tested; 0.14 % in 127 biliary tract cancers tested; 0.13 % in 273 cervix cancers tested; 0.09 % in 603 endometrium cancers tested; 0.08 % in 891 ovary cancers tested; 0.06 % in 589 stomach cancers tested; 0.06 % in 1768 lung cancers tested; 0.05 % in 710 oesophagus cancers tested; 0.05 % in 2082 central nervous system cancers tested; 0.04 % in 441 autonomic ganglia cancers tested; 0.04 % in 1490 breast cancers tested; 0.03 % in 548 urinary tract cancers tested; 0.02 % in 939 prostate cancers tested; 0.02 % in 2009 haematopoietic and lymphoid cancers tested; 0.01 % in 1512 liver cancers tested; 0.01 % in 1360 kidney cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,713 cancer specimens
Comments:
Only 1 deletion and 1 insertion, and no complex mutations are noted on the COSMIC website. About 39% of the point mutations are silent and do not change the amino acid sequence of the protein kinase.

