Nomenclature
Short Name:
PKCi
Full Name:
Protein kinase C, iota/lambda type
Alias:
- APKC-lambda,iota
- MGC26534
- NPKC-iota
- PKC lambda
- PKC-iota
- PKCL; PRKCI, PRKCL
- DXS1179E
- EC 2.7.11.13
- HINT1
- KPCI
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKC
SubFamily:
Iota
Specific Links
Structure
Mol. Mass (Da):
68262
# Amino Acids:
596
# mRNA Isoforms:
1
mRNA Isoforms:
68,262 Da (596 AA; P41743)
4D Structure:
Forms a complex with SQSTM1 and MP2K5 By similarity. Interacts directly with SQSTM1 Probable. Interacts with IKBKB. Interacts with PARD6A, PARD6B and PARD6G. Part of a quaternary complex containing aPKC, PARD3, a PARD6 protein (PARD6A, PARD6B or PARD6G) and a GTPase protein (CDC42 or RAC1). Part of a complex with LLGL1 and PARD6B. Interacts with ADAP1/CENTA1. Interaction with SMG1, through the ZN-finger domain, activates the kinase activity. Interacts with CDK7. Forms a complex with RAB2A and GAPDH involved in recruitment onto the membrane of vesicular tubular clusters (VTCs)
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
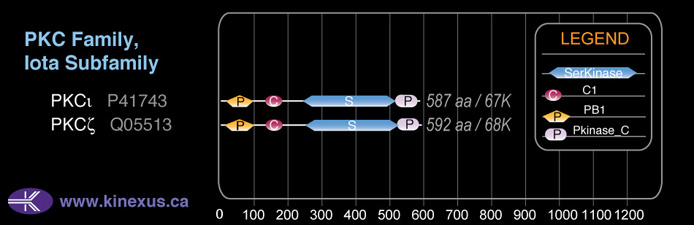
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S7, S8, S11, S19, S44, S46, S222, S223, S226, S246, S247, S248, S411+, S459, S591.
Threonine phosphorylated:
T3, T9, T13, T409+, T410+, T412+, T416+, T557, T564.
Tyrosine phosphorylated:
Y125, Y136+, Y256, Y265, Y271, Y325+, Y334, Y397, Y430, Y584.
Ubiquitinated:
K380, K488.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 34
34
1117
42
1119
 3
3
96
24
99
 5
5
152
3
177
 20
20
645
136
976
 27
27
878
36
776
 10
10
332
119
437
 15
15
482
51
610
 100
100
3245
59
5933
 22
22
718
24
607
 5
5
176
133
227
 2
2
73
38
92
 21
21
668
271
708
 2
2
67
36
63
 3
3
110
21
95
 3
3
112
31
154
 3
3
111
24
124
 9
9
308
434
2010
 6
6
195
18
185
 4
4
119
123
136
 19
19
632
162
704
 4
4
121
28
153
 3
3
89
33
132
 5
5
167
14
109
 5
5
155
18
125
 5
5
171
28
268
 36
36
1153
78
1270
 3
3
104
39
89
 4
4
146
18
136
 4
4
116
18
119
 3
3
89
42
113
 17
17
561
30
563
 52
52
1694
51
3810
 18
18
587
81
1336
 24
24
772
83
675
 2
2
80
48
88
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 66.7
66.7
78.6
100 96.8
96.8
97.5
98 -
-
-
99 -
-
-
97 84.3
84.3
84.9
98.5 -
-
-
- 98.3
98.3
98.8
98.5 72.3
72.3
83.6
99 -
-
-
- 59.9
59.9
60.4
- 71.5
71.5
83
95 89.8
89.8
94.5
93 88.8
88.8
93
90 -
-
-
- 36.4
36.4
54.1
70 68
68
78.2
- 58.1
58.1
74.2
61 63.6
63.6
72.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 24
24
35.7
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PARD6A - Q9NPB6 |
| 2 | CDC42 - P60953 |
| 3 | PARD3 - Q8TEW0 |
| 4 | PARD6B - Q9BYG5 |
| 5 | YWHAZ - P63104 |
| 6 | ANXA1 - P04083 |
| 7 | GAPDH - P04406 |
| 8 | IRAK1 - P51617 |
| 9 | BAD - Q92934 |
| 10 | NULL - Q5BKU6 |
| 11 | MAP2K5 - Q13163 |
| 12 | SMG1 - Q96Q15 |
| 13 | SRC - P12931 |
| 14 | RAB2A - P61019 |
| 15 | RHOQ - P17081 |
Regulation
Activation:
Phosphorylation of Tyr-325, Thr-403 and possible Thr-412 increases phosphotransferase activity. Might be a target for novel lipid activators that are elevated during nutrient-stimulated insulin secretion.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| MARK3 | P27448 | T587 | RGTASRSTFHGQPRE | |
| p70S6Kb (RPS6KB2) | Q9UBS0 | S473 | PPSGTKKSKRGRGRP | + |
| Tau iso8 | P10636-8 | S258 | PDLKNVKSKIGSTEN | |
| Tau iso8 | P10636-8 | S293 | NVQSKCGSKDNIKHV | |
| Tau iso8 | P10636-8 | S324 | KVTSKCGSLGNIHHK | |
| Tau iso8 | P10636-8 | S352 | DFKDRVQSKIGSLDN | |
| AID | Q9GZX7 | T140 | GVQIAIMTFKDYFYC | |
| Bad | Q92934 | S118 | GRELRRMSDEFVDSF | - |
| Bad | Q92934 | S75 | EIRSRHSSYPAGTED | - |
| Bad | Q92934 | S99 | PFRGRSRSAPPNLWA | - |
| Calpain 2 | P17655 | S368 | DGNWRRGSTAGGCRN | |
| Ezrin | P15311 | T567 | QGRDKYKTLRQIRQG | |
| IRAK1 | P51617 | T66 | CERSGQRTASVLWPW | ? |
| MARK2 | Q7KZI7 | T596 | RGVSSRSTFHAGQLR | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
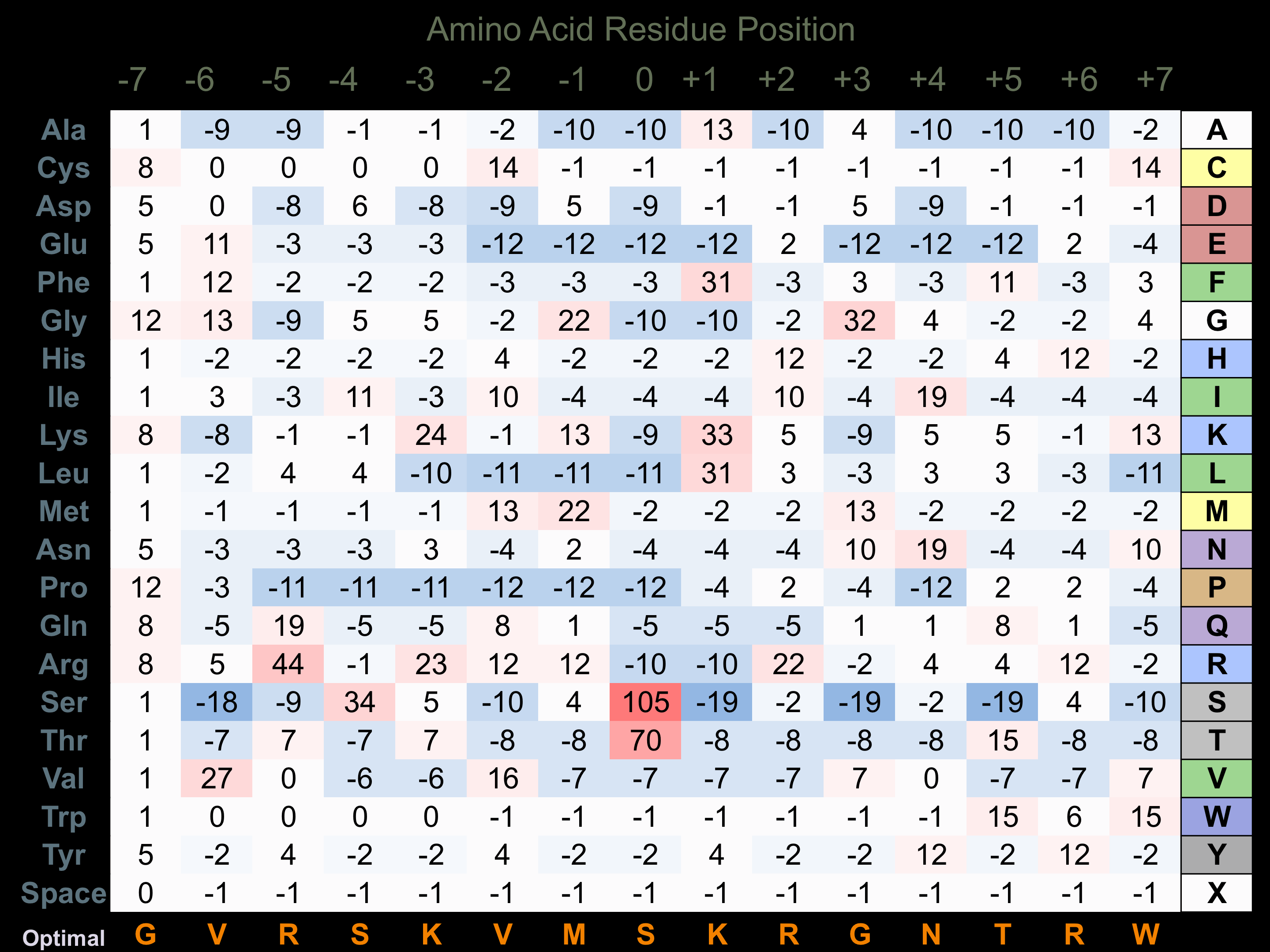
Matrix Type:
Experimentally derived from alignment of 20 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Esophageal squamous cell carcinomas; Lung squamous cell carcinomas; Glioblastoma; Non-small cell lung cancer (NSCLC)
Comments:
PKCi may be an oncoprotein (OP). PKCi activity mediates the resistance of BCR-ABL transformed leukemia cells to apoptotic inducing drugs, thus the protein may play a role in cancer progression and cell survival through the inhibition of apoptosis. In glioblastoma cancer cell lines, PKCi acts downstream of PI3K as an anti-apoptotic factor through phosphorylation and inhibition of the pro-apoptotic factor BAD. In small-cell lung cancer cell lines, PKCi forms a protein complex with PARD6A and ECT2 which functions to positively regulate ECT2 oncogenic activity, thus promoting tumour growth and metastasis. In addition, the transformation and tumour progression of both lung and esophageal squamous cell carcinoma is associated with the amplification of the chromosomal region 3q26, which contains the PKCi, ECT2, PIK3CA, SOX2 and MDS1 genes, thus implicating PKCi in the progression of these cancers. Furthermore, elevated expression of PKCi in esophageal squamous cell carcinoma specimens was significantly correlated with tumour size, metastasis, and advanced stages of cancer progression. Similarly, PKCi over-expression in lung squamous cell carcinoma was associated with the maintenance of a stem-like phenotype of the cancer cells through cooperative action with SOX2 and influence on the sonic-hedgehog (Shh) signalling pathway. Therefore, PKCi is suggested to act as an oncoprotein in the development of several cancer types.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +109, p<0.014); Breast epithelial carcinomas (%CFC= -47, p<0.04); Cervical cancer (%CFC= -49, p<0.003); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +215, p<0.0001); Ovary adenocarcinomas (%CFC= +125, p<0.007); Prostate cancer - primary (%CFC= +95, p<0.0001); Skin squamous cell carcinomas (%CFC= +152, p<0.064). The COSMIC website notes an up-regulated expression score for PKCi in diverse human cancers of 1080, which is 2.3-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 27 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25377 diverse cancer specimens. This rate is only 24 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.74 % in 1093 large intestine cancers tested; 0.35 % in 805 skin cancers tested; 0.17 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R480C (10); R480H (3).
Comments:
Six deletions (all at T276fs*7), 4 insertions (all at T276fs*16), and no complex mutations are noted on the COSMIC website. About 21% of the point mutations are silent and do not change the amino acid sequence of the protein kinase.

