Nomenclature
Short Name:
GRK6
Full Name:
G protein-coupled receptor kinase 6
Alias:
- EC 2.7.11.16
- GPRK6
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
GRK
SubFamily:
GRK
Specific Links
Structure
Mol. Mass (Da):
65,991
# Amino Acids:
576
# mRNA Isoforms:
3
mRNA Isoforms:
67,257 Da (589 AA; P43250-2); 65,991 Da (576 AA; P43250); 64,294 Da (560 AA; P43250-3)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
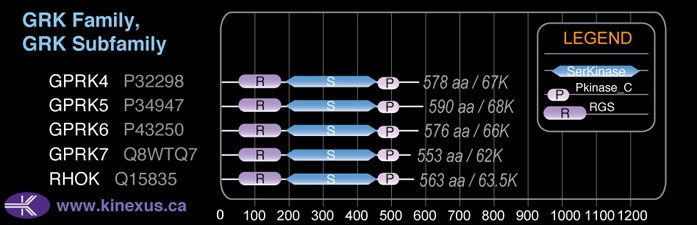
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K235, K240.
Palmitoylated:
C561, C562, C565 (unclear whether palmitoylation is on one or more of C561, C562 and/or C565)RQD CCGNCSDSEE ELPTRL.
Serine phosphorylated:
S78, S111, S484, S557.
Threonine phosphorylated:
T106, T113, T485.
Tyrosine phosphorylated:
Y152, Y161, Y166+, Y351-.
Ubiquitinated:
K139, K240, K343, K358, K402, K426, K487.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 47
47
1174
55
1849
 2
2
55
25
50
 11
11
274
19
639
 16
16
394
170
645
 36
36
886
44
601
 3
3
64
156
156
 7
7
177
55
478
 36
36
907
66
1366
 23
23
582
31
485
 5
5
117
143
297
 3
3
82
50
87
 31
31
781
294
655
 7
7
171
63
162
 2
2
50
18
42
 5
5
135
38
197
 2
2
47
29
36
 11
11
267
145
212
 6
6
152
32
158
 2
2
51
160
60
 21
21
513
215
540
 5
5
118
34
135
 10
10
237
42
293
 12
12
310
31
311
 5
5
117
32
123
 10
10
246
34
235
 34
34
846
100
1335
 9
9
216
66
507
 8
8
198
32
380
 9
9
222
32
714
 22
22
546
56
291
 28
28
702
36
652
 100
100
2491
66
5153
 7
7
172
106
151
 38
38
956
109
748
 15
15
379
61
713
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 94.3
94.3
95.5
98 99.3
99.3
99.8
99 -
-
-
96 -
-
-
96 65.1
65.1
78
98 -
-
-
- 96.7
96.7
98.3
96 96.7
96.7
98.1
96 -
-
-
- 65.9
65.9
69.9
- 73
73
77.1
87 82.1
82.1
89.8
82.5 84.2
84.2
90.6
84 -
-
-
- 50.4
50.4
62.5
- -
-
-
- 53.9
53.9
67
54 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TBXA2R - P21731 |
| 2 | SLC9A3R1 - O14745 |
| 3 | C14orf21 - Q86U38 |
| 4 | ADRB2 - P07550 |
| 5 | RHO - P08100 |
| 6 | BDKRB2 - P30411 |
| 7 | EDNRA - P25101 |
| 8 | RCVRN - P35243 |
| 9 | GIT1 - Q9Y2X7 |
| 10 | FSHR - P23945 |
| 11 | SNCB - Q16143 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
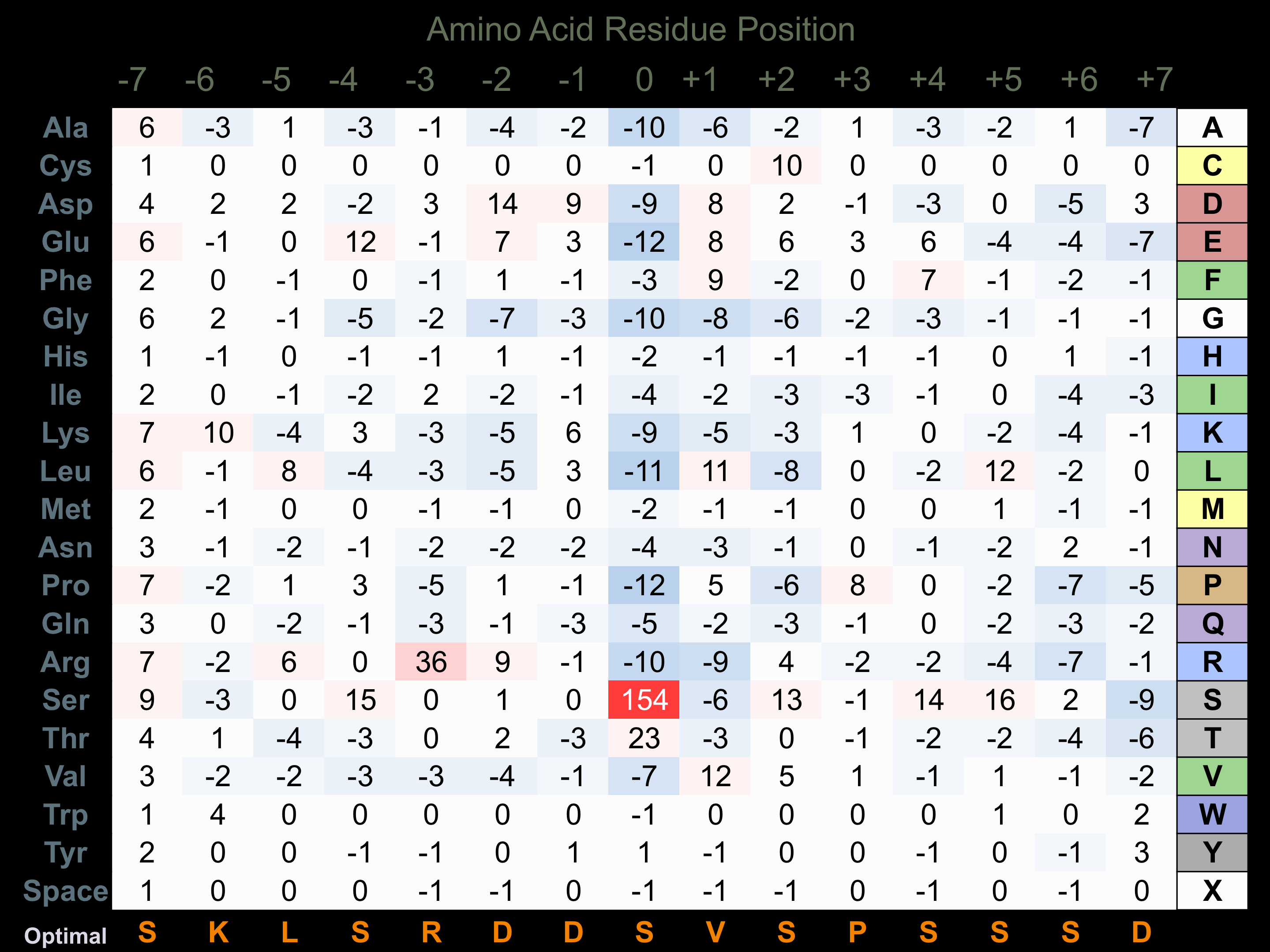
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Staurosporine | IC50 > 50 nM | 5279 | 22037377 | |
| K-252a; Nocardiopsis sp. | IC50 > 150 nM | 3813 | 281948 | 22037377 |
| Gö6976 | IC50 > 250 nM | 3501 | 302449 | 22037377 |
| Ophiocordin | IC50 = 490 nM | 5287736 | 60254 | 20128603 |
| BCP9000906 | IC50 = 500 nM | 5494425 | 21156 | 22037377 |
| IDR E804 | IC50 = 500 nM | 6419764 | 1802727 | 22037377 |
| SU11652 | IC50 = 500 nM | 24906267 | 13485 | 22037377 |
| Syk Inhibitor | IC50 = 500 nM | 6419747 | 104279 | 22037377 |
| A 443654 | IC50 < 1 µM | 10172943 | 379300 | 19465931 |
| MK5108 | IC50 > 1 µM | 24748204 | 20053775 | |
| N-Benzoylstaurosporine | IC50 > 1 µM | 56603681 | 608533 | 22037377 |
| PDK1/Akt/Flt Dual Pathway Inhibitor | IC50 > 1 µM | 5113385 | 599894 | 22037377 |
| PKR Inhibitor | IC50 > 1 µM | 6490494 | 235641 | 22037377 |
| Ro-32-0432 | IC50 > 1 µM | 127757 | 26501 | 22037377 |
| SB218078 | IC50 > 1 µM | 447446 | 289422 | 22037377 |
| SureCN2579964 | IC50 < 2 µM | 24948986 | 22934575 | |
| SureCN2505235 | IC50 < 4 µM | 5353854 | 101797 | 22934575 |
Disease Linkage
General Disease Association:
Blood disorders
Specific Diseases (Non-cancerous):
Whim syndrome
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +119, p<0.006); Brain oligodendrogliomas (%CFC= +114, p<0.02); Breast epithelial carcinomas (%CFC= +74, p<0.004); Breast epithelial cell carcinomas (%CFC= +50, p<0.004); Ovary adenocarcinomas (%CFC= +146, p<0.002); Prostate cancer (%CFC= +57, p<0.011); Skin fibrosarcomas (%CFC= +111, p<0.008); and Skin melanomas - malignant (%CFC= +66, p<0.0001). The COSMIC website notes an up-regulated expression score for GRK6 in diverse human cancers of 415, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 55 for this protein kinase in human cancers was 0.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. Mutations on several sites near the N terminus lead to largely decreased phosphotransferase activity, and mutations on amino acid residues 561, 562, and 565 lead to abolished palmitoylation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24995 diverse cancer specimens. This rate is only -18 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.35 % in 589 stomach cancers tested; 0.26 % in 1270 large intestine cancers tested; 0.2 % in 603 endometrium cancers tested; 0.12 % in 864 skin cancers tested; 0.08 % in 1512 liver cancers tested; 0.05 % in 1822 lung cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,278 cancer specimens
Comments:
Only 4 deletions, and no insertions or complex mutations are noted on the COSMIC website.

