Nomenclature
Short Name:
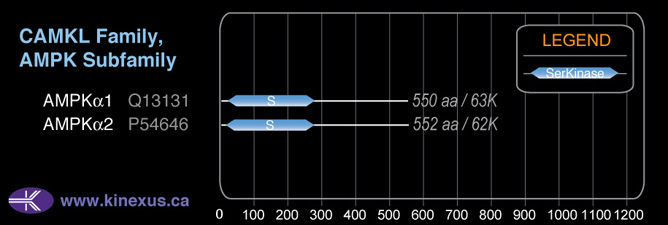
AMPKa2
Full Name:
5'-AMP-activated protein kinase, catalytic alpha-2 chain
Alias:
- AAPK2
- AMPK alpha-2 chain
- EC 2.7.11.1
- Kinase AMPK-alpha2
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMKL
SubFamily:
AMPK
Specific Links
Structure
Mol. Mass (Da):
62,320
# Amino Acids:
552
# mRNA Isoforms:
1
mRNA Isoforms:
62,320 Da (552 AA; P54646)
4D Structure:
Heterotrimer of a catalytic subunit, a beta and a gamma non-catalytic subunits.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 16 | 268 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K69.
Serine phosphorylated:
S161, S165, S173+, S176-, S377, S481, S483, S484, S491, S500, S501, S509, S511, S527, S534.
Threonine phosphorylated:
T172+, T258, T485, T521, T524.
Tyrosine phosphorylated:
Y179-, Y232, Y324.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1276
40
1251
 2
2
23
14
23
 3
3
37
9
58
 19
19
241
165
653
 43
43
550
54
450
 0.8
0.8
10
73
9
 6
6
77
59
240
 23
23
294
49
378
 19
19
239
10
236
 14
14
176
159
194
 2
2
29
32
50
 34
34
435
147
494
 2
2
26
20
49
 2
2
22
9
19
 3
3
42
21
36
 1.2
1.2
15
28
8
 7
7
93
329
963
 4
4
45
16
36
 30
30
388
137
402
 32
32
414
168
321
 3
3
33
28
30
 3
3
32
30
48
 2
2
27
18
39
 3
3
35
16
30
 2
2
25
28
36
 69
69
883
107
1345
 2
2
22
23
22
 4
4
48
16
46
 3
3
39
16
43
 3
3
44
70
51
 27
27
345
18
244
 35
35
450
47
1304
 17
17
214
98
426
 44
44
567
135
526
 3
3
33
74
26
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 76.5
76.5
86.8
100 -
-
-
99 -
-
-
- 73.8
73.8
75.2
99 -
-
-
- 97.6
97.6
98.5
98 98.2
98.2
99.1
98 -
-
-
- 71.1
71.1
81.1
- 27.3
27.3
42.6
94 75.3
75.3
85
94 22.3
22.3
32.7
90 -
-
-
- -
-
-
69 66.1
66.1
77.7
- 53.7
53.7
65.7
63 -
-
-
- -
-
-
- -
-
-
- -
-
-
54 44.9
44.9
62.3
46 33.2
33.2
50.2
52 -
-
-
50
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | STK11 - Q15831 |
| 2 | PRKAG1 - P54619 |
| 3 | PRKAB1 - Q9Y478 |
| 4 | TRIP6 - Q15654 |
| 5 | LEP - P41159 |
| 6 | EEF2K - O00418 |
| 7 | PRKAB2 - O43741 |
| 8 | TP53 - P04637 |
| 9 | PRKAG2 - Q9UGJ0 |
| 10 | PRKAG3 - Q9UGI9 |
| 11 | GYS1 - P13807 |
Regulation
Activation:
Binding of AMP results in allosteric activation, inducing phosphorylation on Thr-172 by STK11 in complex with STE20-related adapter-alpha (STRAD alpha) pseudo kinase and CAB39. Also activated by phosphorylation by CAMKK2 triggered by a rise in intracellular calcium ions, without detectable changes in the AMP/ATP ratio.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
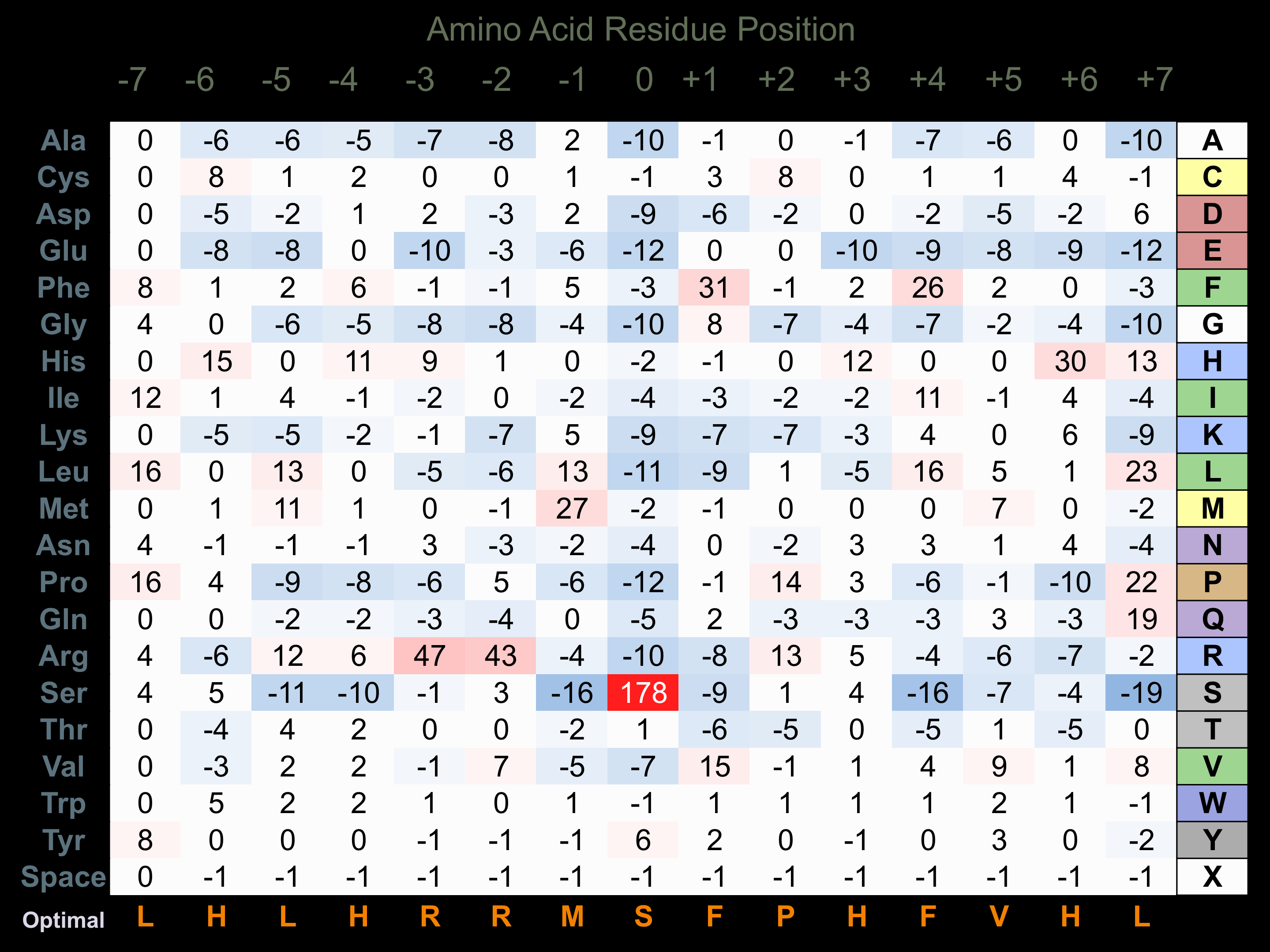
Matrix Type:
Experimentally derived from alignment of 19 known protein substrate phosphosites and 26 peptides phosphorylated by recombinant AMPKa2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| JNJ-28312141 | Kd = 4.1 nM | 22037378 | ||
| Hesperadin | Kd < 10 nM | 10142586 | 514409 | 19035792 |
| Staurosporine | Kd = 12 nM | 5279 | 18183025 | |
| BML-275 | IC50 = 41 nM | 11524144 | 478629 | 20932747 |
| NVP-TAE684 | Kd = 49 nM | 16038120 | 509032 | 22037378 |
| Lestaurtinib | Kd = 56 nM | 126565 | 22037378 | |
| Nintedanib | Kd = 84 nM | 9809715 | 502835 | 22037378 |
| Sunitinib | Kd = 89 nM | 5329102 | 535 | 18183025 |
| SU14813 | Kd = 290 nM | 10138259 | 1721885 | 18183025 |
| Tozasertib | Kd = 350 nM | 5494449 | 572878 | 18183025 |
| Aurora A Inhibitor 23 (DF) | Kd < 400 nM | 21992004 | ||
| KW2449 | Kd = 440 nM | 11427553 | 1908397 | 22037378 |
| N-Benzoylstaurosporine | Kd = 460 nM | 56603681 | 608533 | 18183025 |
| PHA-665752 | Kd = 770 nM | 10461815 | 450786 | 22037378 |
| Aurora A Inhibitor 1 (DF) | Kd < 800 nM | 21992004 | ||
| Dovitinib | Kd = 820 nM | 57336746 | 18183025 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Foretinib | Kd = 1.2 µM | 42642645 | 1230609 | 22037378 |
| JNJ-7706621 | Kd = 1.8 µM | 5330790 | 191003 | 18183025 |
| TG101348 | Kd = 4.7 µM | 16722836 | 1287853 | 22037378 |
Disease Linkage
General Disease Association:
Cancer, cellular metabolism disregulation; Arrhythmia; Unbalanced sex hormones
Specific Diseases (Non-cancerous):
Ischemia; Wolff-Parkinson-White syndrome; Aromatase deficiency; Phosphorylase kinase deficiency
Comments:
Ischemia occurs when there is a lack of oxygen and nutrient supply to tissues which often includes the brain, spinal cord, and heart. >Wolff-Parkinson-White Syndrome (WPW) is a relatively rare disease where sufferers have heart arrhythmia. Tissues affected in Wolff-Parkinson-White Syndrome include the heart, testes, and spinal cord. >Aromatase deficiency leads to reduced estrogen (female sex hormone) and increased testosterone (male sex hormone), with affected tissues being the bone, ovary, and placenta.
Specific Cancer Types:
Peutz-Jeghers syndrome
Comments:
Defects in PRKAA2 are linked to Peutz-Jeghers Syndrome, which is a relatively rare autosomal dominant disease where benign polyps develop on the GI tract. Sufferers of Peutz-Jeghers Syndrome are at increased risk for other cancers and affected tissue can also include colon, small intestine, and skin. A phosphomimetic mutation that is observed is T172D.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -47, p<0.001); and Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +420, p<0.002). The COSMIC website notes an up-regulated expression score for AMPKa2 in diverse human cancers of 400, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 2 for this protein kinase in human cancers was 97% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. A phosphomimetic mutation in PRKAA2 that is observed corresponds to T172D.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 25279 diverse cancer specimens. This rate is a modest 1.59-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.01 % in 805 skin cancers tested; 0.6 % in 1119 large intestine cancers tested; 0.31 % in 589 stomach cancers tested; 0.17 % in 1755 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R293C (4); R293H (2).
Comments:
Ten deletions, 1 insertion and no complex mutations are noted on the COSMIC website.

