Nomenclature
Short Name:
PDGFRA
Full Name:
Alpha platelet-derived growth factor receptor
Alias:
- Alpha platelet-derived growth factor receptor precursor
- PDGFR2
- PDGFRa
- PDGFR-alpha
- PDGF-R-alpha; PGFRA; Platelet-derived growth factor, alpha-receptor; Rhe-PDGFRA
- AI115593
- CD140a
- EC 2.7.10.1
- Kinase PDGFR-alpha
- MGC74795
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
PDGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
122,670
# Amino Acids:
1089
# mRNA Isoforms:
3
mRNA Isoforms:
122,670 Da (1089 AA; P16234); 82,809 Da (743 AA; P16234-3); 24,023 Da (218 AA; P16234-2)
4D Structure:
Homodimer, and heterodimer with PDGFRB. Interacts with the SH2 domain of SHB via phosphorylated Tyr-720 By similarity. Interacts with the SH2 domain of SHF via phosphorylated Tyr-720.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
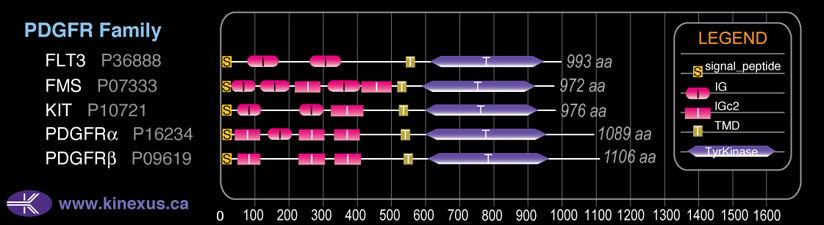
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
K378.
N-GlcNAcylated:
N42, N76, N103, N179, N353, N359,N458, N468.
Serine phosphorylated:
S566, S584, S716, S752, S755, S760, S767, S847+, S935, S1016, S1041, S1042, S1048, S1080, S1081, S1087.
Threonine phosphorylated:
T717, T739, T740, T992.
Tyrosine phosphorylated:
Y572, Y574, Y582, Y613, Y720, Y731, Y742, Y754, Y762, Y768, Y849+, Y926, Y944, Y958, Y962, Y988, Y993, Y1018.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 60
60
1158
48
1354
 7
7
144
27
217
 9
9
173
24
179
 15
15
293
177
545
 36
36
698
41
699
 4
4
80
126
169
 24
24
468
69
632
 19
19
359
75
516
 32
32
610
24
625
 8
8
164
170
356
 5
5
98
64
202
 27
27
513
339
592
 4
4
70
57
82
 18
18
353
25
505
 9
9
166
33
434
 6
6
114
30
216
 6
6
115
486
1607
 5
5
93
41
215
 5
5
88
163
135
 29
29
561
185
648
 4
4
86
54
90
 9
9
172
59
259
 9
9
173
43
160
 5
5
90
42
208
 3
3
60
53
77
 27
27
518
109
691
 2
2
48
60
56
 5
5
89
39
79
 8
8
157
42
156
 2
2
39
42
29
 10
10
190
30
234
 100
100
1935
62
5096
 9
9
169
123
422
 32
32
618
114
604
 4
4
70
61
69
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 45
45
63.3
97 44.3
44.3
63.6
99 -
-
-
94 -
-
-
98 95.7
95.7
98
96 -
-
-
- 91.8
91.8
96.2
92 90.9
90.9
96
91 -
-
-
- 62.8
62.8
70.2
- 80.3
80.3
89.2
81 75.6
75.6
85.1
76 60.9
60.9
75.5
64 61.6
61.6
75.3
- -
-
-
- -
-
-
- -
-
-
42 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
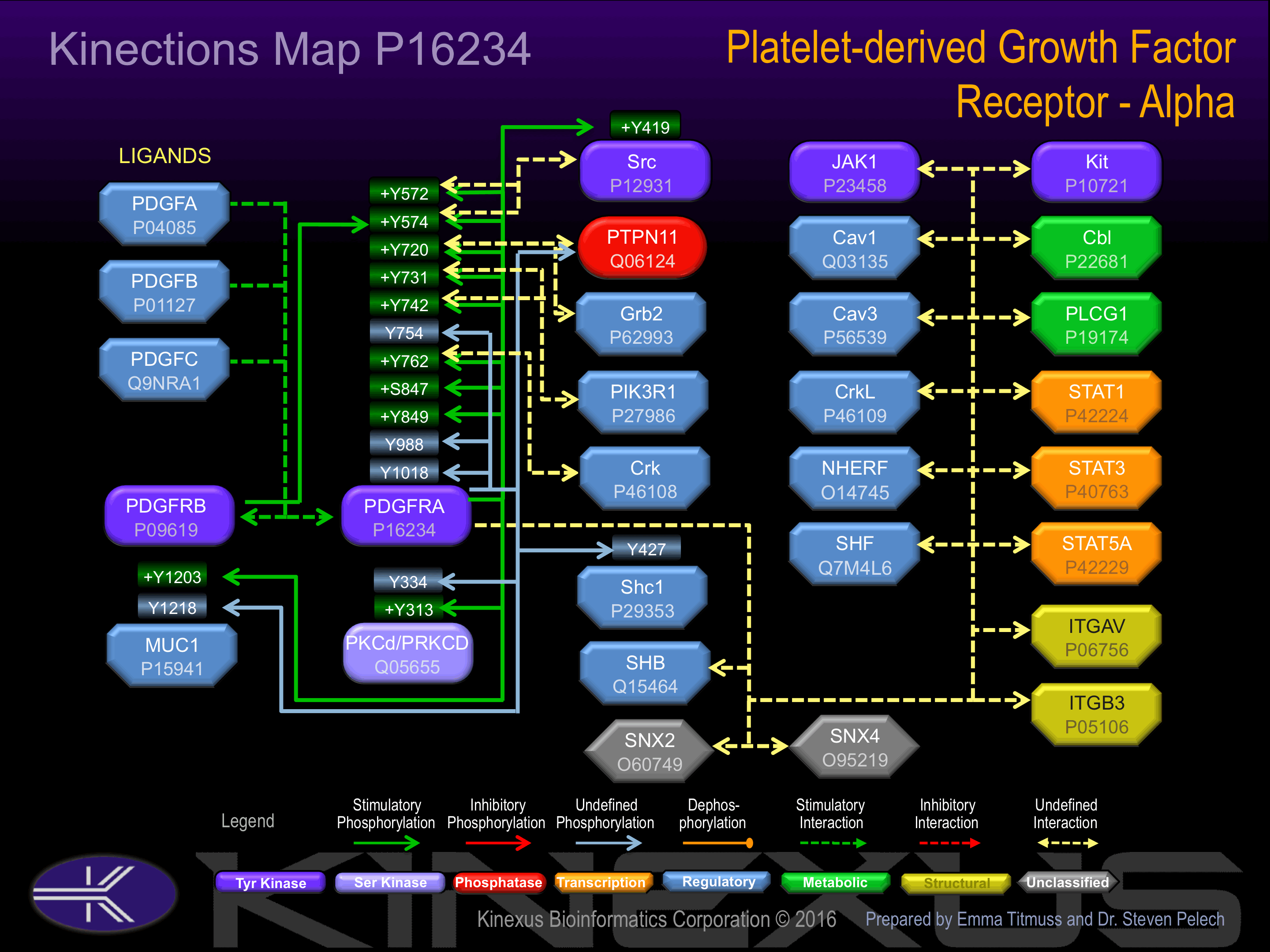
Activated by binding platelet-derived growth factor (PDGF), which induces dimerization and autophosphorylation. Autophosphorylation of Tyr-572 and Tyr-574 induces interaction with Src. Autophosphorylation of Tyr-720 and Tyr-754 induces interaction with protein-tyrosine phosphatase SHP2. Autophosphorylation of Tyr-731 and Tyr-742 induces interaction with PIK3R1. Autophosphorylation of Tyr-762 induces interaction with CDK6 and CrkL. Autophosphorylation of Tyr-988 and Tyr-1018 induces interaction with PLCg1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PDGFRA | P16234 | Y572 | ISPDGHEYIYVDPMQ | ? |
| PDGFRA | P16234 | Y574 | PDGHEYIYVDPMQLP | ? |
| PDGFRA | P16234 | Y720 | ADESTRSYVILSFEN | ? |
| PDGFRA | P16234 | Y731 | SFENNGDYMDMKQAD | ? |
| PDGFRA | P16234 | Y742 | KQADTTQYVPMLERK | ? |
| PDGFRB | P09619 | Y754 | ERKEVSKYSDIQRSL | |
| PDGFRA | P16234 | Y754 | ERKEVSKYSDIQRSL | |
| PDGFRA | P16234 | Y762 | SDIQRSLYDRPASYK | ? |
| PDGFRA | P16234 | Y988 | RVDSDNAYIGVTYKN | |
| PDGFRA | P16234 | Y1018 | RLSADSGYIIPLPDI |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| MUC1 | P15941 | Y1203 | IFPARDTYHPMSEYP | |
| MUC1 | P15941 | Y1218 | TYHTHGRYVPPSSTD | |
| PDGFRA | P16234 | Y1018 | RLSADSGYIIPLPDI | |
| PDGFRA | P16234 | Y572 | ISPDGHEYIYVDPMQ | ? |
| PDGFRA | P16234 | Y574 | PDGHEYIYVDPMQLP | ? |
| PDGFRA | P16234 | Y720 | ADESTRSYVILSFEN | ? |
| PDGFRA | P16234 | Y731 | SFENNGDYMDMKQAD | ? |
| PDGFRA | P16234 | Y742 | KQADTTQYVPMLERK | ? |
| PDGFRA | P16234 | Y754 | ERKEVSKYSDIQRSL | |
| PDGFRA | P16234 | Y762 | SDIQRSLYDRPASYK | ? |
| PDGFRA | P16234 | Y988 | RVDSDNAYIGVTYKN | |
| PKCd (PRKCD) | Q05655 | Y313 | SSEPVGIYQGFEKKT | + |
| PKCd (PRKCD) | Q05655 | Y334 | MQDNSGTYGKIWEGS | ? |
| Shc1 | P29353 | Y427 | ELFDDPSYVNVQNLD | ? |
| Src | P12931 | Y419 | RLIEDNEYTARQGAK | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
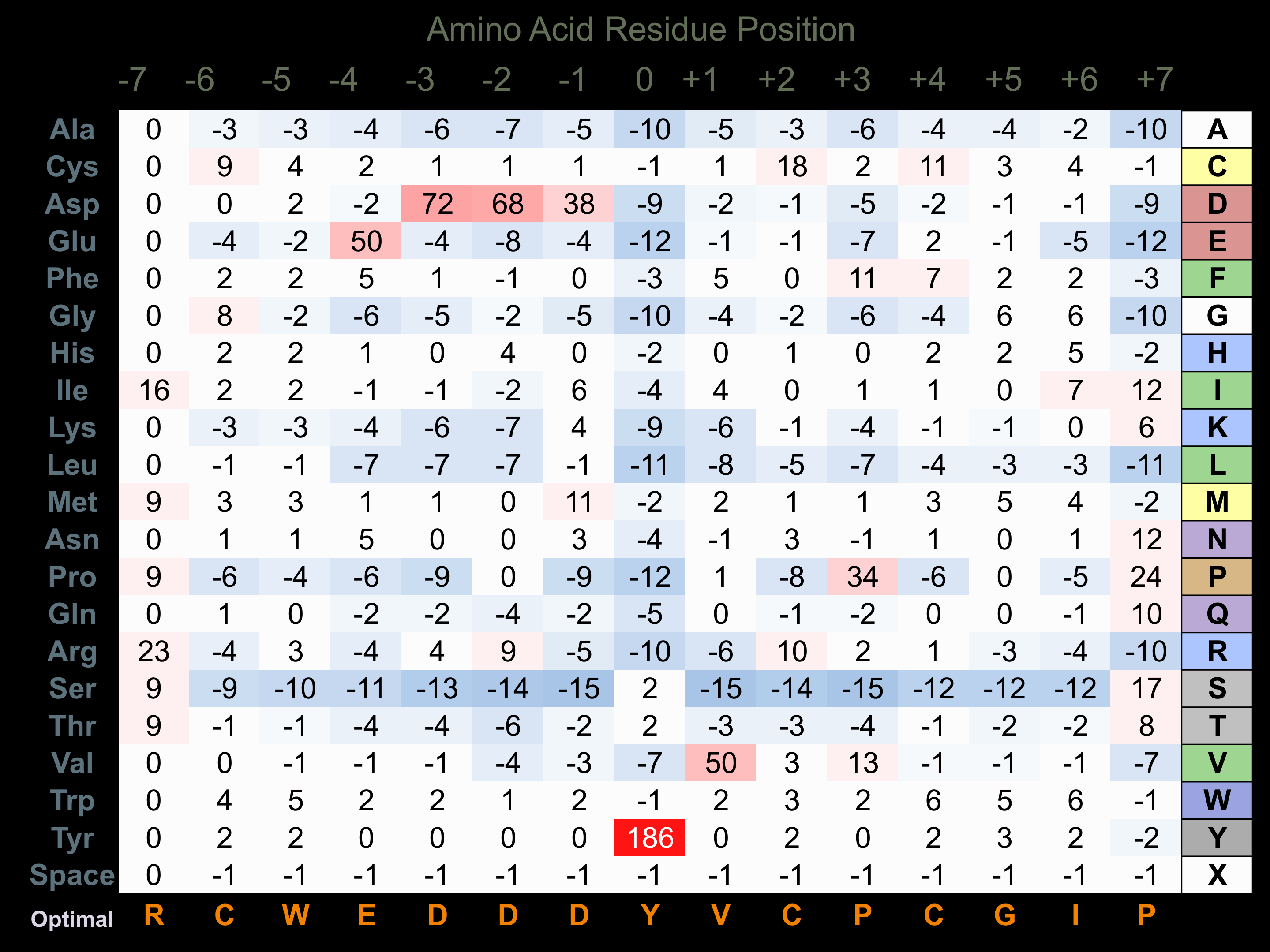
Matrix Type:
Experimentally derived from alignment of 9 known protein substrate phosphosites and 101 peptides phosphorylated by recombinant PDGFRA in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, developmental, and blood vessel disorders
Specific Diseases (Non-cancerous):
Neural tube defects (NTDS); Hereditary hemorrhagic telangiectasia (HHT); Pulmonary vein stenosis
Comments:
Neural Tube Defects (NTDS) is a rare condition affecting neonates, often during the first month of pregnancy, where the spine does not close properly (spina bifida). NTDS can lead to at least partial paralysis of the legs. Hereditary Hemorrhagic Telangiectasia (HHT) is a blood vessel disorder which is characterized by excessive bleeding caused by improperly developed blood vessels. HHT can manifest in the brain, lungs, liver, or intestines. Pulmonary Vein Stenosis arises from the thickening of pulmonary vein walls and narrowing of the lumen, leading to obstructed blood flow.
Specific Cancer Types:
Myeloid neoplasm associated with PDGFRA rearrangement; Gastrointestinal stromal tumours (GANT, GIST); PDGFRA-associated chronic eosinophilic leukemias; Hypereosinophilic syndrome (HES); Neurofibromatosis (NF1); Piebaldism (PBT); Fibrosarcomas; Mastocytosis; Desmoid tumours; Oligodendrogliomas; Leiomyosarcomas; Neurofibromas; Carney triad; Medulloblastomas (MDB); Small cell neuroendocrine carcinomas; Adenosquamous carcinomas; Large cell carcinomas; Fibrosarcomas of bone; Cervical adenosquamous carcinomas; Osteosarcomas; Astrocytomas; Juvenile myelomonocytic leukemias (JMML); Chondromas; Sarcomatoid renal cell carcinomas; Perineuriomas; Choroid plexus carcinomas; Rosai-Dorfman disease (RDD); Oligoastrocytomas (MOA); Pediatric osteosarcomas; Endomyocardial fibrosis (EMF); Mixed gliomas; Mesenchymal cell neoplasms; Gastric leiomyosarcoma; Hypereosinophilic syndrome, Idiopathic, Resistant to Imatinib; Reticular perineuriomas; Gastrointestinal stromal tumours, somatic; leukemias, acute lymphoblastic 3 (B-ALL); Gastrointestinal stromal tumours, familial
Comments:
PDGFRA is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. PDGFRA is linked to Gastrointestinal Stromal tumours (GANT, GIST), which are soft tissue sarcomas growing from the stomach, intestines, or rectum. A constitutively active PDGFRA can be induced with a V561D, N659K, D842V, or D842Y, mutation, or with the deletion of 842-845, or deletion of 845-848. Pdgfra-Associated Chronic Eosinophilic Leukemia is a rare blood cancer where the eosinophil count is elevated. Hypereosinophilic Syndrome (HES) is a rare blood disorder with increased eosinophil levels and can affect lungs, kidneys, heart, and nervous system. Neurofibromatosis (NF1) is a rare cancer where tumours grow underneath the skin or along the nerves. NF1 typically manifests through changes in skin and deformed bones. In the rare condition Piebaldism (PBT) there is a lack of pigment producing cells (melanocytes) in various areas around the skin and hair. Individuals suffering from PBT suffer from increased risks of skin cancer in the non-pigmented patches. The Fibrosarcoma disorder induces malignant tumours arising from fibrous connective tissues and can affect bone, breast, and colon tissues. Mastocytosis is a rare condition arising from an overproduction of mast cells in the body. Desmoid tumour is a rare form of cancer arising from connective tissues, and can be aggressive in nature. Oligodendroglioma can affect brain, spinal cord, and endothelial tissues and is a cancer arising from oligodendrocytes. Leiomyosarcoma is a rare sarcoma of the smooth muscle cells which can also affect the lung, liver, blood vessels, or other soft tissues. Carney Triad is related to the gastrointestinal stromal tumour and paranglioma disorders. Carney Triad is often characterized by mediastinal anomalies, and arrhythmia. Medulloblastoma (MDB) is a rare, highly malignant brain tumour. Large Cell Carcinoma is characterized by a large cytoplasm component with a cell that has rounded or polygonal-shaped features. Osteosarcoma accounts for the most bone marrow cancers in youth, but can also be associated with lung and breast tissue. Juvenile Myelomonocytic Leukemia (JMML) is a rare chronic blood leukemia that mostly affects young (under 4 years) children. Chondroma is a rare, benign tumour arising from cartilage. Chondroma can either grow within the bone (enchondroma), or outward from the bone (ecchondroma). Perineurioma is rare and relates to the granular cell tumour and neurofibromatosis disorders. The rare cancer, Choroid Plexus Carcinoma, has a relation to the activation (via transcription ligand) ESR1 SP pathway and to the pathway involving retinoblastoma protein regulation. Rosai-Dorfman Disease (RDD) is characterized by the increased presence of white blood cells in the body's lymph nodes, though more prevalently the neck. Oligoastrocytoma (MOA) is a rare brain tumour where the glial cells called oligodendrocytes and astrocytes form the tumour. Endomyocardial Fibrosis (EMF) is related to the condition hypereosinophilic syndrome where eosinophil count is consistently elevated. EMF can affect heart, liver, and brain tissues. Hypereosinophilic Syndrome, Idiopathic, Resistant to Imatinib is a rare cancer of the blood, and related pathways include HTLV-I infection and MAPK signalling. Reticular Perineurioma has a relation to the perineurioma and myelofibrosis conditions. Leukemia, Acute Lymphoblastic 3 (B-ALL) is a rare cancer which is related to the gefitinib and the transcription factors in neurogenesis pathways. Gastrointestinal Stromal Tumor, Familial is a rare condition related to the gastrointestinal stromal tumour and myelofibrosis conditions.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain oligodendrogliomas (%CFC= -79, p<0.05); Breast epithelial carcinomas (%CFC= -61, p<0.053); Colon mucosal cell adenomas (%CFC= -54, p<0.002); Ovary adenocarcinomas (%CFC= -76, p<0.026); Skin melanomas - malignant (%CFC= -88, p<0.019); Uterine fibroids (%CFC= -57, p<0.052). The COSMIC website notes an up-regulated expression score for PDGFRA in diverse human cancers of 407, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. Y574F and Y572F mutations, in conjunction, can lead to impaired Src-family proteins interacting with PDGFRA. PTPN11 and GRB2 interaction with PDGFRA can be prevented with a Y720F mutation. Activation of phosphatidylinositol 3-kinase is inhibited with either a Y731F or Y742F mutation. Interaction of PDGFRA with CRK can be impaired with a Y762F mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.36 % in 45368 diverse cancer specimens. This rate is 4.8-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.03 % in 10775 soft tissue cancers tested; 0.5 % in 928 stomach cancers tested; 0.47 % in 1715 large intestine cancers tested; 0.38 % in 1662 skin cancers tested; 0.31 % in 2467 lung cancers tested; 0.25 % in 747 endometrium cancers tested; 0.16 % in 2703 central nervous system cancers tested; 0.13 % in 3447 haematopoietic and lymphoid cancers tested; 0.05 % in 2818 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: D842V (493); V561D (61); N659K (27); T674I (7).
Comments:
The PDGFRA gene undergoes extensive mutations in restricted regions: 53 of the deletion mutations are located at I843 in the kinase catalytic domain, 8 insertion mutations and 63 complex mutations at S566 are located just before the kinase catalytic domain as noted on the COSMIC website.