Nomenclature
Short Name:
PDGFRB
Full Name:
Beta platelet-derived growth factor receptor
Alias:
- CD140b
- PDGFRb
- PDGFR-beta
- PDGF-R-beta
- Platelet-derived growth factor receptor, beta polypeptide
- EC 2.7.10.1
- JTK12
- Kinase PDGFR-beta
- PDGFR
- PDGFR1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
PDGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
123,968
# Amino Acids:
1106
# mRNA Isoforms:
2
mRNA Isoforms:
123,968 Da (1106 AA; P09619); 37,412 Da (336 AA; P09619-2)
4D Structure:
Homodimer, and heterodimer with PDGFRA. Interacts with APS. The autophosphorylated form interacts directly with SHB and with PIK3C2B, maybe indirectly. Interacts with GRB10
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
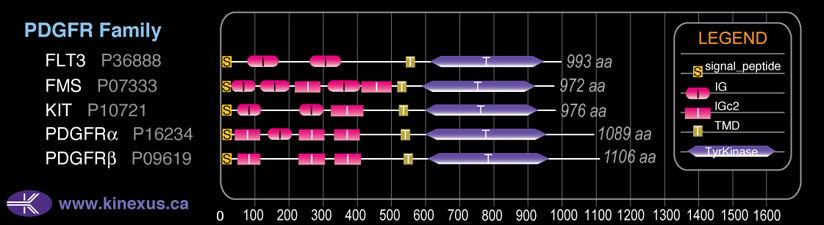
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N45, N89, N103, N215, N230, N292, N307, N354, N371, N468, N479.
Serine phosphorylated:
S212, S213, S608+, S712, S717, S855+, S930, S1104.
Threonine phosphorylated:
T605.
Tyrosine phosphorylated:
Y562, Y579+, Y581+, Y678, Y683, Y686+, Y692, Y716+, Y740+, Y751+, Y763, Y771, Y775, Y778, Y857+, Y921, Y934+, Y970+, Y1009, Y1021.
Ubiquitinated:
K707.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-

 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 96
96
96.4
98 96
96
96.4
98 -
-
-
91 -
-
-
98 98.3
98.3
99.1
90 44.4
44.4
62.9
- -
-
-
86 85.5
85.5
92.3
86 85.8
85.8
92.3
- 47.8
47.8
59.1
- 47.8
47.8
59.1
68.5 43.6
43.6
60.9
- 45.5
45.5
63
53 43.1
43.1
60.2
- 31.7
31.7
47.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PLCG1 - P19174 |
| 2 | PIK3R1 - P27986 |
| 3 | PTPN11 - Q06124 |
| 4 | NCK1 - P16333 |
| 5 | GRB2 - P62993 |
| 6 | PDGFB - P01127 |
| 7 | NCK2 - O43639 |
| 8 | PTPN1 - P18031 |
| 9 | CRK - P46108 |
| 10 | SHC1 - P29353 |
| 11 | SLC9A3R1 - O14745 |
| 12 | PTEN - P60484 |
| 13 | SLC9A3R2 - Q15599 |
| 14 | RAF1 - P04049 |
| 15 | SYNGAP1 - Q96PV0 |
Regulation
Activation:
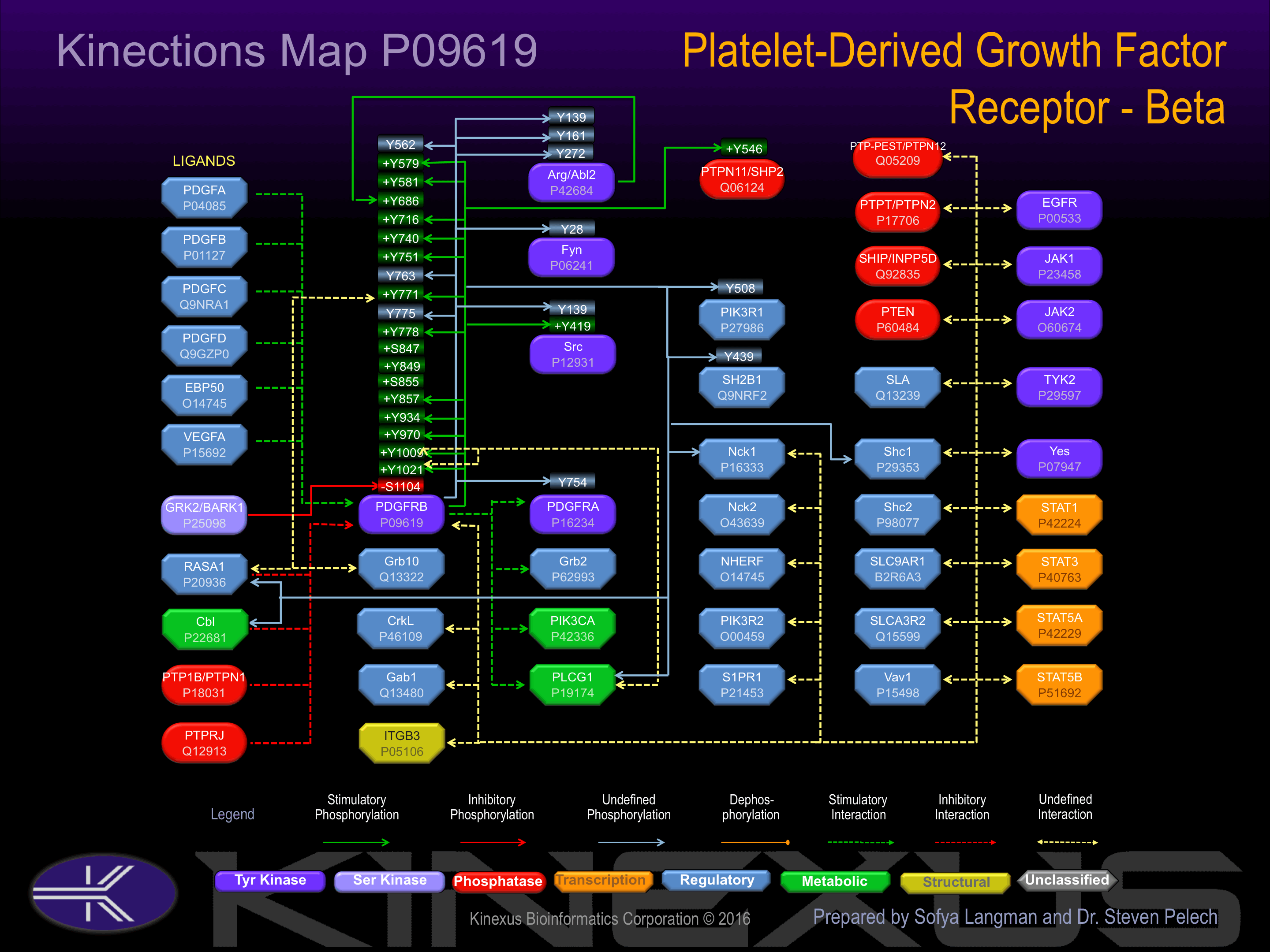
Activated by binding platelet-derived growth factor, which induces dimerization and autophosphorylation. Autophosphorylation of Tyr-1021 induces interaction with PLCg1. Autophosphorylation of Tyr-579 increases phosphotransferase activity and induces interaction with PIK3R1, PLCg1, RasGAP, Shc1 and Src. Autophosphorylation of Tyr-581 increases phosphotransferase activity and induces interaction with PIK3R1, PLCg1, RasGAP, and Src. Autophosphorylation of Tyr-716 increases phosphotransferase activity and induces interaction with Grb2 and Grb7. Autophosphorylation of Tyr-740, Tyr-751 increases phosphotransferase activity and induces interaction with PIK3C2A, PIK3CA, PIK3R1, and Shc1. Autophosphorylation of Tyr-763 induces interaction with protein-tyrosine phosphatase SHP2. Autophosphorylation of Tyr-771 induces interaction with RasGAP and Shc1. Autophosphorylation of Tyr-775 induces interaction with Grb7.
Inhibition:
Phosphorylation of Tyr-1009 induces interaction with Cbl, PLCg1, SH3KBP1 and SHP2, and inhibits binding to PIK3R1 and RasGAP.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PDGFRB | P09619 | Y562 | LWQKKPRYEIRWKVI | |
| PDGFRB | P09619 | Y579 | VSSDGHEYIYVDPMQ | + |
| PDGFRB | P09619 | Y581 | SDGHEYIYVDPMQLP | + |
| ARG | P42684 | Y686 | IITEYCRYGDLVDYL | + |
| PDGFRB | P09619 | Y716 | RPPSAELYSNALPVG | + |
| PDGFRB | P09619 | Y740 | TGESDGGYMDMSKDE | + |
| PDGFRB | P09619 | Y751 | SKDESVDYVPMLDMK | + |
| PDGFRB | P09619 | Y763 | DMKGDVKYADIESSN | ? |
| PDGFRB | P09619 | Y771 | ADIESSNYMAPYDNY | ? |
| PDGFRB | P09619 | Y775 | SSNYMAPYDNYVPSA | ? |
| PDGFRB | P09619 | Y778 | YMAPYDNYVPSAPER | |
| PDGFRB | P09619 | Y857 | DIMRDSNYISKGSTF | + |
| PDGFRB | P09619 | Y1009 | LDTSSVLYTAVQPNE | ? |
| PDGFRB | P09619 | Y1021 | PNEGDNDYIIPLPDP | ? |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Arg (Abl2) | P42684 | Y139 | EKLRVLGYNQNGEWS | |
| Arg (Abl2) | P42684 | Y161 | QGWVPSNYITPVNSL | |
| Arg (Abl2) | P42684 | Y272 | KCNKPTVYGVSPIHD | |
| Fyn | P06241 | Y28 | SLNQSSGYRYGTDPT | |
| PDGFRA | P16234 | Y754 | ERKEVSKYSDIQRSL | |
| PDGFRB | P09619 | Y1009 | LDTSSVLYTAVQPNE | ? |
| PDGFRB | P09619 | Y1021 | PNEGDNDYIIPLPDP | ? |
| PDGFRB | P09619 | Y562 | LWQKKPRYEIRWKVI | |
| PDGFRB | P09619 | Y579 | VSSDGHEYIYVDPMQ | + |
| PDGFRB | P09619 | Y581 | SDGHEYIYVDPMQLP | + |
| PDGFRB | P09619 | Y716 | RPPSAELYSNALPVG | + |
| PDGFRB | P09619 | Y740 | TGESDGGYMDMSKDE | + |
| PDGFRB | P09619 | Y751 | SKDESVDYVPMLDMK | + |
| PDGFRB | P09619 | Y763 | DMKGDVKYADIESSN | ? |
| PDGFRB | P09619 | Y771 | ADIESSNYMAPYDNY | ? |
| PDGFRB | P09619 | Y775 | SSNYMAPYDNYVPSA | ? |
| PDGFRB | P09619 | Y778 | YMAPYDNYVPSAPER | |
| PDGFRB | P09619 | Y857 | DIMRDSNYISKGSTF | + |
| PIK3R1 | P27986 | Y508 | QERYSKEYIEKFKRE | |
| PTPN11 (SHP2) | Q06124 | Y546 | SKRKGHEYTNIKYSL | + |
| SH2-Bb | Q9NRF2 | Y439 | DRLSQGAYGGLSDRP | |
| Src | P12931 | Y139 | TGYIPSNYVAPSDSI | |
| Src | P12931 | Y419 | RLIEDNEYTARQGAK | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
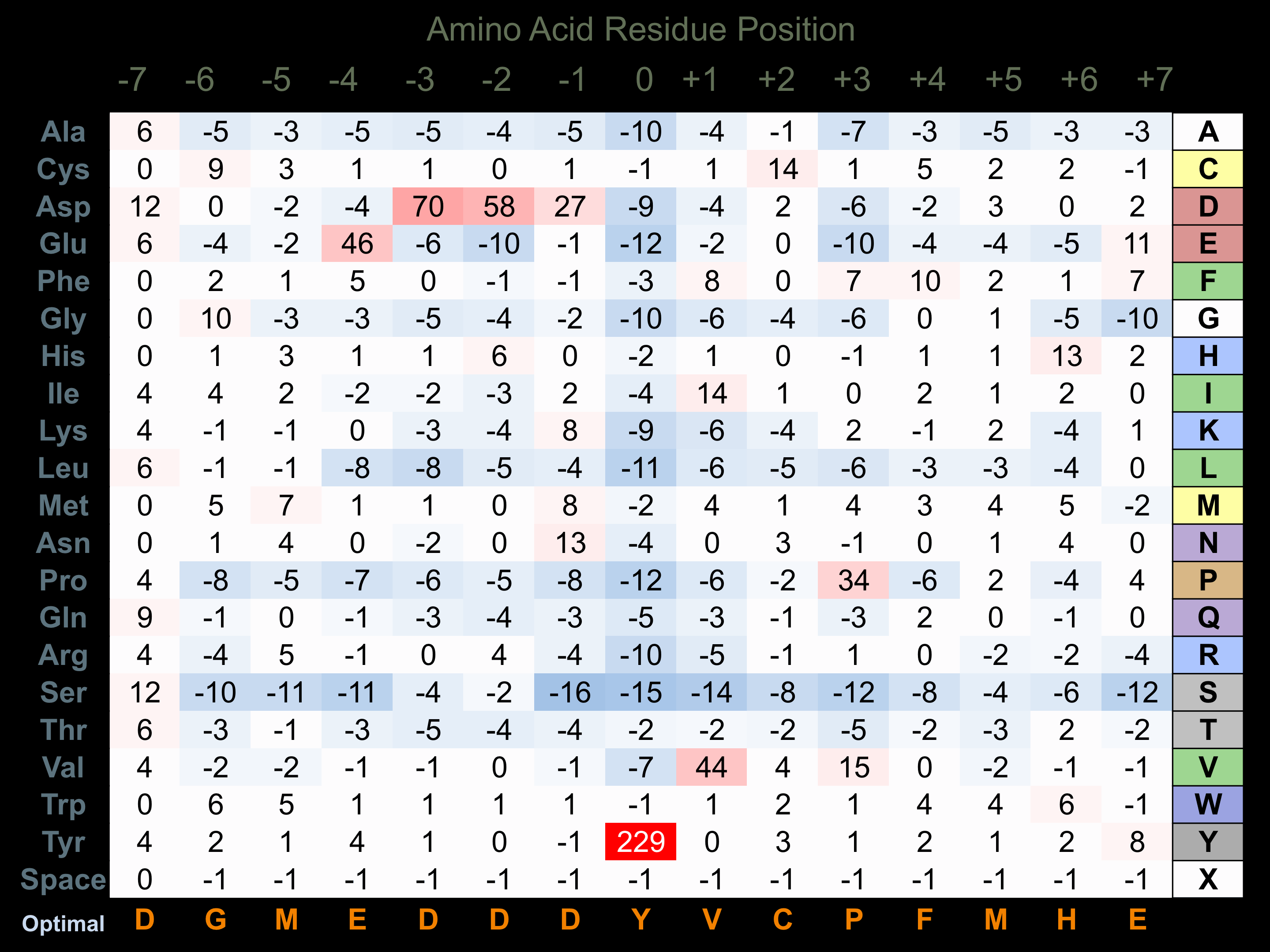
Matrix Type:
Experimentally derived from alignment of 26 known protein substrate phosphosites and 64 peptides phosphorylated by recombinant PDGFRB in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, blood, immune, cardiovascular, and bone disorders
Specific Diseases (Non-cancerous):
Eosinophilia; Chronic myeloproliferative disease; Polycythemia vera; Mastocytosis; Hypereosinophilic syndrome; Basal Ganglia Calcification, Idiopathic, 4; Myeloproliferative disorder with Eosinophilia; Infantile Myofibromatosis; Pigmented Villonodular Synovitis; Synovitis; Diabetic Angiopathy; Villonodular Synovitis; Tendinosis; Primary Familial Brain Calcification; Basal Ganglia Calcification, Idiopathic, 1; Hypereosinophilic syndrome, Idiopathic, Resistant to Imatinib; Unclassified Chronic myeloproliferative disease; Primary Familial Brain Calcification 4
Comments:
L658P and R987W substitutions in PDGFRB are associated with Basal ganglia calcification, idiopathic, 4, which is a form of basal ganglia calcification and can show a wide range of neuropsychiatric symptoms. Heterozygous missense R5611 mutations in this gene have been identified with infantile Myofibromatosis 1.
Specific Cancer Types:
Myeloid neoplasm associated with PDGFRB rearrangement; PDGFRB-associated chronic eosinophilic leukemias; Myeloproliferative disorder; Chronic myelomonocytic leukemias (CMML); Gastrointestinal stromal tumours (GIST); Dermatofibrosarcoma protuberans (DFSP); Dermatofibrosarcoma; desmoid tumours; Gliosarcomas; Pilocytic astrocytomas (PA); Uterine sarcomas; Myofibromatosis, infantile, 1; Astrocytomas; brain cancer; Solitary fibrous tumours (SFT); Choroid plexus carcinomas (CPC)
Comments:
PDGFRB is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. R561C and P660T mutations in PDGFRB are associated with myofibromatosis, infantile 1, which is a rare mesenchymal disorder featured by the development of benign tumours in multiple sites. A translocation between chromosomes 5 and 12 that fuses this gene to that of the translocation, results in chronic myeloproliferative disorder with eosinophilia.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +48, p<0.016); Breast epithelial carcinomas (%CFC= +141, p<0.025); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -54, p<0.004); Colorectal adenocarcinomas (early onset) (%CFC= +138, p<0.001); Gastric cancer (%CFC= +66, p<0.003); Oral squamous cell carcinomas (OSCC) (%CFC= +138, p<0.013); Uterine fibroids (%CFC= +81, p<0.015); and Vulvar intraepithelial neoplasia (%CFC= -57, p<0.032). The COSMIC website notes an up-regulated expression score for PDGFRB in diverse human cancers of 330, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 3 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25820 diverse cancer specimens. This rate is only 22 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.52 % in 805 skin cancers tested; 0.39 % in 1152 large intestine cancers tested; 0.34 % in 629 stomach cancers tested; 0.2 % in 500 urinary tract cancers tested; 0.18 % in 602 endometrium cancers tested; 0.13 % in 1942 lung cancers tested; 0.06 % in 1544 breast cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,915 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.