Nomenclature
Short Name:
PKCb
Full Name:
Protein kinase C, beta type
Alias:
- EC 2.7.11.13
- PKC-beta
- PKC-II
- PRKCB
- PRKCB1; PRKCB2; Protein kinase C, beta type
- Kinase PKC-beta
- KPCB
- MGC41878
- PKC II
- PKC-B
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKC
SubFamily:
Alpha
Specific Links
Structure
Mol. Mass (Da):
76869
# Amino Acids:
671
# mRNA Isoforms:
2
mRNA Isoforms:
77,012 Da (673 AA; P05771-2); 76,869 Da (671 AA; P05771)
4D Structure:
Interacts with PDK1 By similarity. Interacts in vitro with PRKCBP1. Interacts with PHLPP1 and PHLPP2; both proteins mediate its dephosphorylation. Interacts with KDM1A/LSD1, PKN1 and ANDR
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
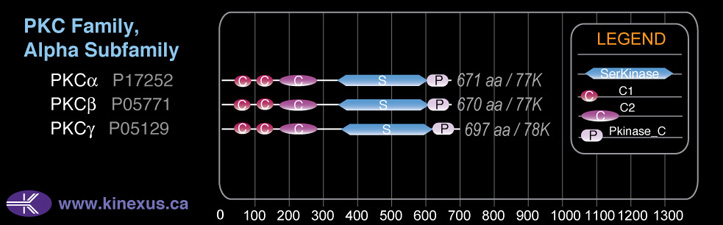
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
A2, K197.
Serine phosphorylated:
S11, S16, S51, S206, S241, S311, S352, S408, S476, S659, S661-.
Threonine phosphorylated:
T17, T48, T134, T226, T314, T324, T340, T364, T412, T497+, T498+, T500+, T504-, T635+, T642+, T644.
Tyrosine phosphorylated:
Y123, Y195, Y368, Y417, Y422, Y507-, Y515, Y662.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 54
54
1345
56
1053
 1.1
1.1
27
21
23
 6
6
140
18
88
 35
35
885
213
876
 18
18
451
62
330
 2
2
52
116
143
 10
10
258
83
445
 100
100
2494
67
6025
 15
15
363
17
265
 3
3
71
222
73
 2
2
56
47
61
 22
22
549
262
628
 13
13
319
40
169
 1
1
24
14
29
 1.4
1.4
35
26
44
 0.8
0.8
19
36
19
 3
3
76
535
721
 3
3
85
29
98
 2
2
46
184
37
 19
19
464
221
413
 5
5
114
41
79
 11
11
265
45
231
 3
3
75
44
68
 3
3
85
29
75
 9
9
223
41
182
 67
67
1659
137
4119
 9
9
226
43
145
 3
3
67
29
65
 2
2
56
29
53
 10
10
253
84
305
 59
59
1467
36
643
 55
55
1369
62
2167
 11
11
265
159
444
 27
27
668
171
624
 7
7
187
87
125
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
96 99.8
99.8
99.8
100 -
-
-
98 -
-
-
100 90.1
90.1
91.8
99 -
-
-
- 94.6
94.6
96.2
99 98.8
98.8
99.2
98 -
-
-
- 79.1
79.1
87.6
- 86.1
86.1
89.7
92 76.8
76.8
85.2
91 32.6
32.6
50.5
88 -
-
-
- 66.5
66.5
80.7
70 70.8
70.8
81.2
- 65.8
65.8
78.8
67 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
39
- 26
26
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BTK - Q06187 |
| 2 | ADRBK1 - P25098 |
| 3 | LMNB1 - P20700 |
| 4 | RASGRP3 - Q8IV61 |
| 5 | NCF1B - A6NI72 |
| 6 | EIF6 - P56537 |
| 7 | ANXA2P2 - A6NMY6 |
| 8 | MARCKS - P29966 |
| 9 | GAP43 - P17677 |
| 10 | PDPK1 - O15530 |
| 11 | NCAM1 - P13591 |
| 12 | RB1 - P06400 |
| 13 | IFNAR2 - P48551 |
| 14 | SYT6 - Q5T7P8 |
| 15 | PDPK2 - Q6A1A2 |
Regulation
Activation:
Activated by calcium, diacylglycerol, and acidic phospholipids such as phosphatidylserine. Phosphorylation of Thr-635 and Thr-642 is critical for phosphotransferase activity.
Inhibition:
Phosphorylation of Ser-661 inhibits membrane binding and causes release into cytoplasm.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKCb | P05771 | S16 | PPSEGEESTVRFARK | |
| PKCb | P05771 | T17 | PSEGEESTVRFARKG | |
| PKCb | P05771 | T314 | RAKISQGTKVPEEKT | |
| PKCb | P05771 | T324 | PEEKTTNTVSKFDNN | |
| PKCb | P05771 | T500 | WDGVTTKTFCGTPDY | + |
| PKCa | P17252 | T500 | WDGVTTKTFCGTPDY | + |
| BLVRA | P53004 | T500 | WDGVTTKTFCGTPDY | + |
| PKCb | P05771 | T635 | SNFDKEFTRQPVELT | + |
| PKCb | P05771 | T642 | TRQPVELTPTDKLFI | + |
| PKCb | P05771 | S659 | QSEFEGFSFVNSEFL | |
| PKCb | P05771 | S659 | QSEFEGFSFVNSEFL | |
| PKCb | P05771 | S661 | QNEFAGFSYTNPEFV | - |
| SYK | P43405 | Y662 | NEFAGFSYTNPEFVI |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AID | Q9GZX7 | T140 | GVQIAIMTFKDYFYC | |
| Akt1 (PKBa) | P31749 | S473 | RPHFPQFSYSASGTA | + |
| Akt1 (PKBa) | P31749 | T308 | KDGATMKTFCGTPEY | + |
| ANXA2 | P07355 | S25 | TPPSAYGSVKAYTNF | |
| ATF2 | P15336 | S121 | LATPIIRSKIEEPSV | |
| ATF2 | P15336 | S340 | TPQTQSTSGRRRRAA | |
| ATF2 | P15336 | S367 | ERNRAAASRCRQKRK | |
| ATP1A1 | P05023 | S16 | KYEPAAVSEQGDKKG | |
| Btk | Q06187 | S180 | GSLKPGSSHRKTKKP | - |
| C5aR | P21730 | S334 | SVVRESKSFTRSTVD | |
| CARD11 | Q9BXL7 | S559 | QPPRSRSSIMSITAE | |
| CARD11 | Q9BXL7 | S644 | NLMFRKFSLERPFRP | |
| CARD11 | Q9BXL7 | S645 | LERPFRPSVTSVGHV | |
| CD5 | P06127 | T434 | MSFHRNHTATVRSHA | |
| CD5 | P06127 | T436 | FHRNHTATVRSHAEN | |
| CFLAR | O15519 | S193 | LQAAIQKSLKDPSNN | |
| ChAT | P28329 | S464 | LLKHMTQSSRKLIRA | |
| ChAT | P28329 | S465 | LKHMTQSSRKLIRAD | |
| ChAT | P28329 | S558 | VPTYESASIRRFQEG | |
| ChAT | P28329 | S594 | HKAAVPASEKLLLLK | |
| ChAT | P28329 | T373 | TVLVKDSTNRDSLDM | |
| Cx43 | P17302 | S367 | QRPSSRASSRASSRP | |
| DAB2 | P98082 | S24 | QAAPKAPSKKEKKKG | |
| EGFR | P00533 | T678 | RHIVRKRTLRRLLQE | - |
| EPB41 | P11171 | S521 | QAQTRQASALIDRPA | |
| GAP43 | P17677 | S41 | AATKIQASFRGHITR | |
| Gg(12) | Q9UBI6 | S2 | _______SSKTASTN | |
| GSK3a | P49840 | S21 | SGRARTSSFAEPGGG | - |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| HMGA1 | P17096 | S44 | PGTALVGSQKEPSEV | |
| HMGA1 | P17096 | S64 | PRGRPKGSKNKGAAK | |
| IRS1 | P35568 | S323 | MVGGKPGSFRVRASS | |
| IRS1 | P35568 | S441 | SPCDFRSSFRSVTPD | |
| ITGB2 | P05107 | S745 | FEKEKLKSQWNNDNP | |
| ITGB2 | P05107 | T758 | NPLFKSATTTVMNPK | |
| Kv3.4 | Q03721 | S15 | SSYRGRKSGNKPPSK | |
| Kv3.4 | Q03721 | S21 | KSGNKPPSKTCLKEE | |
| Kv3.4 | Q03721 | S8 | MISSVCVSSYRGRKS | |
| Kv3.4 | Q03721 | S9 | ISSVCVSSYRGRKSG | |
| mGluR5 | P41594 | S840 | VRSAFTTSTVVRMHV | |
| NOX5 | Q96PH1 | S544 | RSVTMRKSQRSSKGS | + |
| NOX5 | Q96PH1 | T540 | KRLSRSVTMRKSQRS | + |
| OSBPL9 | Q96SU4 | S287 | PNTVPEFSYSSSEDE | |
| p47phox | P14598 | S303 | RGAPPRRSSIRNAHS | + |
| p47phox | P14598 | S304 | GAPPRRSSIRNAHSI | + |
| p47phox | P14598 | S315 | AHSIHQRSRKRISQD | ? |
| p70S6Kb (RPS6KB2) | Q9UBS0 | S473 | PPSGTKKSKRGRGRP | + |
| PEBP1 | P30086 | S153 | RGKFKVASFRKKYEL | |
| PKCa (PRKCA) | P17252 | T497 | MDGVTTRTFCGTPDY | + |
| PKCb (PRKCB) | P05771 | S16 | PPSEGEESTVRFARK | |
| PKCb (PRKCB) | P05771 | S661 | QNEFAGFSYTNPEFV | - |
| PKCb (PRKCB) | P05771 | T17 | PSEGEESTVRFARKG | |
| PKCb (PRKCB) | P05771 | T314 | RAKISQGTKVPEEKT | |
| PKCb (PRKCB) | P05771 | T324 | PEEKTTNTVSKFDNN | |
| PKCb (PRKCB) | P05771 | T500 | WDGVTTKTFCGTPDY | + |
| PKCb (PRKCB) | P05771 | T635 | SNFDKEFTRQPVELT | + |
| PKCb (PRKCB) | P05771 | T642 | TRQPVELTPTDKLFI | + |
| PKCb2 (PRKCB) iso2 | P05771 | S659 | QSEFEGFSFVNSEFL | |
| PLCB3 | Q01970 | S1105 | LDRKRHNSISEAKMR | - |
| PTPN11 (SHP2) | Q06124 | S580 | CAEMREDSARVYENV | |
| PTPN11 (SHP2) | Q06124 | S595 | GLMQQQKSFR_____ | |
| RasGRP3 | Q8IV61 | T133 | YDWMRRVTQRKKISK | |
| RBCK1 | Q9BYM8 | S137 | LLSARNTSLNPQELQ | |
| RBCK1 | Q9BYM8 | S272 | HVQLDQRSLVLNTEP | |
| RBCK1 | Q9BYM8 | S287 | AECPVCYSVLAPGEA | |
| RBCK1 | Q9BYM8 | T161 | DLGFKDLTLQPRGPL | |
| RBCK1 | Q9BYM8 | T209 | CTFINKPTRPGCEMC | |
| RBCK1 | Q9BYM8 | T277 | QRSLVLNTEPAECPV | |
| SNAP25 | P60880 | S187 | RIMEKADSNKTRIDE | |
| STMN1 | P16949 | S25 | QAFELILSPRSKESV | |
| STMN1 | P16949 | S38 | SVPEFPLSPPKKKDL | |
| Tau iso8 | P10636-8 | S258 | PDLKNVKSKIGSTEN | |
| Tau iso8 | P10636-8 | S293 | NVQSKCGSKDNIKHV | |
| Tau iso8 | P10636-8 | S324 | KVTSKCGSLGNIHHK | |
| Tau iso8 | P10636-8 | S352 | DFKDRVQSKIGSLDN | |
| TH | P07101 | S61 | SYTPTPRSPRFIGRR | |
| TNNI3 | P19429 | S22 | PAPIRRRSSNYRAYA | |
| TNNI3 | P19429 | S23 | APIRRRSSNYRAYAT | |
| TOP2A | P11388 | T1343 | FSDFDEKTDDEDFVP | |
| TYR | P14679 | S523 | MEKEDYHSLYQSHL_ | |
| TYR | P14679 | S527 | DYHSLYQSHL_____ | |
| VDR | P11473 | S51 | CKGFFRRSMKRKALF | + |
| ATF2 | P15336 | S121 | LATPIIRSKIEEPSV | |
| C5aR | P21730 | S334 | SVVRESKSFTRSTVD | |
| CACNA1B | Q00975 | S1757 | FEMLKHMSPPLGLGK | |
| CACNA1B | Q00975 | S2111 | RRQERGRSQERRQPS | |
| CACNA1B | Q00975 | S424 | KRAATKKSRNDLIHA | |
| CACNA1B | Q00975 | T421 | DVLKRAATKKSRNDL | |
| CACNA1D | Q01668 | S81 | PPPVGSLSQRKRQQY | |
| ChAT | P28329 | S464 | LLKHMTQSSRKLIRA | |
| ChAT | P28329 | S465 | LKHMTQSSRKLIRAD | |
| ChAT | P28329 | S558 | VPTYESASIRRFQEG | |
| ChAT | P28329 | S594 | HKAAVPASEKLLLLK | |
| ChAT | P28329 | T373 | TVLVKDSTNRDSLDM | |
| DAB2 | P98082 | S24 | QAAPKAPSKKEKKKG | |
| GABRB1 | P18505 | S434 | GRIRRRASQLKVKIP | |
| GABRB3 | P28472 | S432 | KTHLRRRSSQLKIKI | |
| GABRB3 | P28472 | S433 | THLRRRSSQLKIKIP | |
| Ghrelin | Q9UBU3 | S41 | RVQQRKESKKPPAKL | |
| GSK3a | P49840 | S21 | SGRARTSSFAEPGGG | - |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| hnRNP P2 | P35637 | S257 | GRGGMGGSDRGGFNK | |
| ITGB2 | P05107 | S745 | FEKEKLKSQWNNDNP | |
| ITGB2 | P05107 | T758 | NPLFKSATTTVMNPK | |
| JNK2 (MAPK9) | P45984 | S129 | ELDHERMSYLLYQML | + |
| KIR3DL1 | P43629 | S415 | QRKITRPSQRPKTPP | |
| Lamin B1 | P20700 | S395 | LKLSPSPSSRVTVSR | |
| Lamin B1 | P20700 | S405 | VTVSRASSSRSVRTT | |
| M-CK | P06732 | S128 | LDPNYVLSSRVRTGR | |
| mGluR5 | P41594 | S840 | VRSAFTTSTVVRMHV | |
| MYH9 | P35579 | S1915 | AMNREVSSLKNKLRR | |
| p47phox | P14598 | S303 | RGAPPRRSSIRNAHS | + |
| p47phox | P14598 | S304 | GAPPRRSSIRNAHSI | + |
| p47phox | P14598 | S315 | AHSIHQRSRKRLSQD | ? |
| p47phox | P14598 | S320 | QRSRKRLSQDAYRRN | ? |
| p47phox | P14598 | S328 | QDAYRRNSVRFLQQR | + |
| p47phox | P14598 | S359 | EERQTQRSKPQPAVP | + |
| p47phox | P14598 | S370 | PAVPPRPSADLILNR | + |
| p47phox | P14598 | S379 | DLILNRCSESTKRKL | ? |
| p70S6Kb (RPS6KB2) | Q9UBS0 | S473 | PPSGTKKSKRGRGRP | + |
| p73 | O15350 | S388 | VPQPLVDSYRQQQQL | |
| PEBP1 | P30086 | S153 | RGKFKVASFRKKYEL | |
| PKCb2 (PRKCB) iso2 | P05771 | S659 | QSEFEGFSFVNSEFL | |
| PPARA | Q07869 | S179 | RFGRMPRSEKAKLKA | ? |
| PPARA | Q07869 | S230 | VKARVILSGKASNNP | |
| PTPN11 (SHP2) | Q06124 | S580 | CAEMREDSARVYENV | |
| PTPN11 (SHP2) | Q06124 | S595 | GLMQQQKSFR_____ | |
| SYT6 | Q5T7P8 | T283 | DRKCKLQTRVHRKTL | |
| SYT6 | Q5T7P8 | T417 | RRLKKKKTTIKKNTL | |
| SYT6 | Q5T7P8 | T418 | RLKKKKTTIKKNTLN | |
| Tau iso8 | P10636-8 | S258 | PDLKNVKSKIGSTEN | |
| Tau iso8 | P10636-8 | S293 | NVQSKCGSKDNIKHV | |
| Tau iso8 | P10636-8 | S324 | KVTSKCGSLGNIHHK | |
| Tau iso8 | P10636-8 | S352 | DFKDRVQSKIGSLDN | |
| TNNI3 | P19429 | S22 | PAPIRRRSSNYRAYA | |
| TNNI3 | P19429 | S23 | APIRRRSSNYRAYAT | |
| TNNI3 | P19429 | S41 | AKKKSKISASRKLQL | |
| TNNI3 | P19429 | S43 | KKSKISASRKLQLKT | |
| TNNI3 | P19429 | T142 | RGKFKRPTLRRVRIS |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
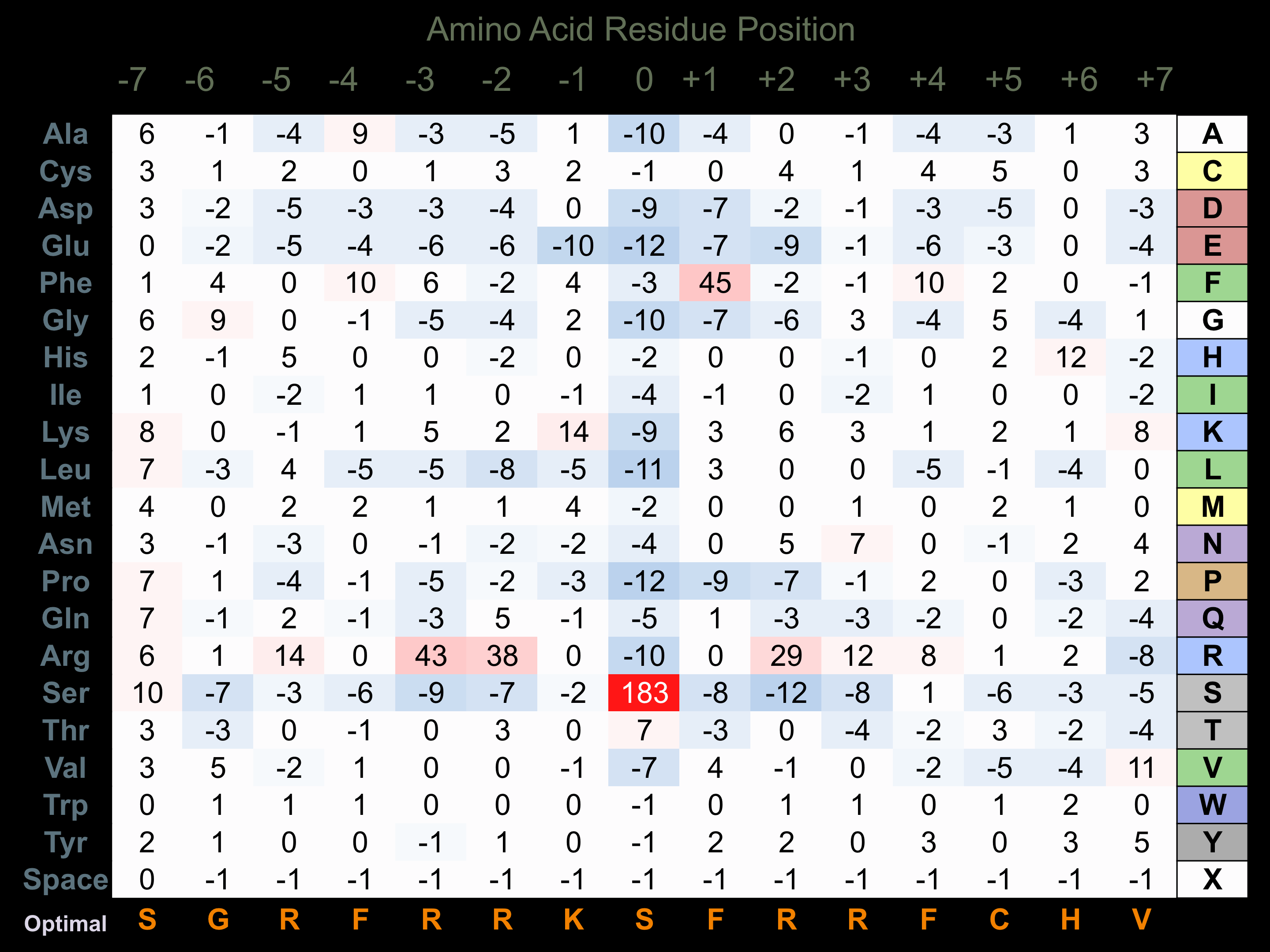
Matrix Type:
Experimentally derived from alignment of 91 known protein substrate phosphosites and 40 peptides phosphorylated by recombinant PKCb in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, eye disorders
Specific Diseases (Non-cancerous):
Diabetic macular edema
Comments:
Diabetic Macular Edema is an eye disease arising from diabetes.
Specific Cancer Types:
Prostate cancer
Comments:
PKCb may be an oncoprotein (OP). A higher Gleason Score significantly associated with increased PKCb expression in prostate cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer stage 2A (%CFC= -56, p<0.103); Classical Hodgkin lymphomas (%CFC= -79, p<0.003); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +48, p<0.043); Colorectal adenocarcinomas (early onset) (%CFC= +111, p<0.004); Large B-cell lymphomas (%CFC= -62, p<0.02); Malignant pleural mesotheliomas (MPM) tumours (%CFC= -49, p<0.007); Oral squamous cell carcinomas (OSCC) (%CFC= +164, p<0.0001); T-cell prolymphocytic leukemia (%CFC= -45, p<0.0001); Uterine fibroids (%CFC= +83, p<0.005); and Uterine leiomyomas from fibroids (%CFC= +115, p<0.031). The COSMIC website notes an up-regulated expression score for PKCb in diverse human cancers of 270, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 25596 diverse cancer specimens. This rate is 1.64-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.49 % in 10 peritoneum cancers tested; 0.64 % in 864 skin cancers tested; 0.49 % in 1270 large intestine cancers tested; 0.3 % in 589 stomach cancers tested; 0.22 % in 603 endometrium cancers tested; 0.21 % in 710 oesophagus cancers tested; 0.17 % in 1512 liver cancers tested; 0.16 % in 382 soft tissue cancers tested; 0.15 % in 1956 lung cancers tested; 0.11 % in 548 urinary tract cancers tested; 0.1 % in 891 ovary cancers tested; 0.09 % in 1490 breast cancers tested; 0.09 % in 1360 kidney cancers tested; 0.07 % in 2103 central nervous system cancers tested; 0.06 % in 939 prostate cancers tested; 0.06 % in 2009 haematopoietic and lymphoid cancers tested; 0.05 % in 273 cervix cancers tested; 0.05 % in 1467 pancreas cancers tested; 0.04 % in 1078 upper aerodigestive tract cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: A509T (5).
Comments:
Only 2 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website. About 30% of the point mutations are silent and do not change the amino acid sequence of the protein kinase.

