Nomenclature
Short Name:
SYK
Full Name:
Tyrosine-protein kinase SYK
Alias:
- DKFZp313N1010
- EC 2.7.10.2
- Spleen tyrosine kinase
- SYK
- FLJ25043
- FLJ37489
- Kinase Syk
- KSYK
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Syk
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
72,066
# Amino Acids:
635
# mRNA Isoforms:
2
mRNA Isoforms:
72,066 Da (635 AA; P43405); 69,510 Da (612 AA; P43405-2)
4D Structure:
Interacts with RHOH, GAB2 and with NFAM1 (phosphorylated form). Interacts (via its SH2 domains) with CD79A (via its phosphorylated ITAM domain); the interaction stimulates SYK autophosphorylation and activation By similarity. Interacts (phosphorylated form) with SLA; the interaction may link it to CBL, leading to its destruction. Interacts with Epstein-Barr virus LMP2A. Interacts with FCRL3. Interacts (via the second SH2 domain) with USP25 (via C-terminus).
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
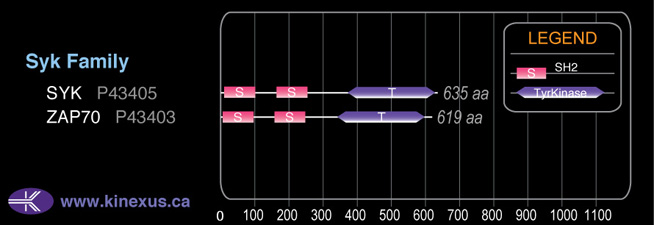
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K105, K116, K394, K402, K571.
Serine phosphorylated:
S178, S202, S295, S297, S316, S319, S350, S579.
Threonine phosphorylated:
T73-, T161, T345, T504, T530.
Tyrosine phosphorylated:
Y28, Y47, Y74, Y91, Y131-, Y203, Y244, Y296, Y323+, Y348+, Y352+, Y431, Y507, Y525+, Y526+, Y539, Y546, Y568, Y629, Y630, Y631.
Ubiquitinated:
K60, K509, K517.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1393
41
1286
 1.1
1.1
15
19
11
 23
23
320
14
308
 21
21
286
154
1008
 56
56
777
43
547
 4
4
49
94
48
 25
25
346
59
536
 92
92
1287
51
3097
 47
47
648
17
543
 10
10
133
162
161
 6
6
87
38
160
 41
41
576
235
628
 31
31
430
36
688
 0.5
0.5
7
11
7
 9
9
127
37
173
 2
2
28
26
23
 10
10
140
360
1684
 19
19
268
25
354
 3
3
41
137
57
 45
45
627
165
552
 16
16
218
33
339
 33
33
465
39
620
 20
20
278
24
374
 17
17
234
26
665
 35
35
487
35
641
 67
67
928
100
852
 35
35
489
38
680
 15
15
207
26
306
 10
10
141
25
200
 7
7
100
56
93
 26
26
363
24
369
 49
49
687
46
658
 22
22
312
96
331
 90
90
1249
109
901
 11
11
148
61
203
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 85.5
85.5
85.7
95 99.1
99.1
99.5
99 -
-
-
93 -
-
-
92 91.2
91.2
94.4
93 -
-
-
- 92.1
92.1
94.5
93 91
91
93.7
92 -
-
-
- -
-
-
- 77.5
77.5
87.4
81 67.7
67.7
79.1
- 66
66
77.5
72 -
-
-
- 25.6
25.6
39.9
- -
-
-
- -
-
-
- 26.5
26.5
40.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CBL - P22681 |
| 2 | VAV1 - P15498 |
| 3 | FCGR2A - P12318 |
| 4 | PTPN6 - P29350 |
| 5 | CBLB - Q13191 |
| 6 | CD79B - P40259 |
| 7 | BLNK - Q8WV28 |
| 8 | POU2AF1 - Q16633 |
| 9 | CD22 - P20273 |
| 10 | HCLS1 - P14317 |
| 11 | DBNL - Q9UJU6 |
| 12 | SLC4A1 - P02730 |
| 13 | CSF2RB - P32927 |
| 14 | TUBA4A - P68366 |
| 15 | MS4A2 - Q01362 |
Regulation
Activation:
Phosphorylation of Tyr-348 and Tyr-352 enhances the binding, phosphorylation and activation of phospholipase C-gamma and the early phase of Ca(2+) mobilization via a phosphoinositide 3-kinase-independent pathway. Phosphorylation of Tyr-525 and Tyr-526 increases phosphotransferase activity.
Inhibition:
Phosphorylation of Tyr-131 inhibits interaction with Bcr. Phosphorylation of Tyr-323 negatively regulates B cell Ag receptor signaling and strongly dampens the Ca(2+) signal.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| SYK | P43405 | Y131 | KENLIREYVKQTWNL | - |
| SYK | P43405 | Y323 | STVSFNPYEPELAPW | + |
| LCK | P06239 | Y323 | STVSFNPYEPELAPW | + |
| LYN | P07948 | Y323 | STVSFNPYEPELAPW | + |
| SYK | P43405 | Y348 | LPMDTEVYESPYADP | + |
| LCK | P06239 | Y348 | LPMDTEVYESPYADP | + |
| LYN | P07948 | Y348 | LPMDTEVYESPYADP | + |
| SYK | P43405 | Y352 | TEVYESPYADPEEIR | + |
| LCK | P06239 | Y352 | TEVYESPYADPEEIR | + |
| LYN | P07948 | Y352 | TEVYESPYADPEEIR | + |
| SYK | P43405 | Y525 | ALRADENYYKAQTHG | + |
| SYK | P43405 | Y526 | LRADENYYKAQTHGK | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| BLNK | Q8WV28 | Y178 | LLEDEADYVVPVEDN | + |
| BLNK | Q8WV28 | Y189 | VEDNDENYIHPTESS | + |
| BLNK | Q8WV28 | Y72 | SDDFDSDYENPDEHS | + |
| BLNK | Q8WV28 | Y84 | EHSDSEMYVMPAEEN | + |
| BLNK | Q8WV28 | Y96 | EENADDSYEPPPVEQ | + |
| Btk | Q06187 | Y551 | RYVLDDEYTSSVGSK | + |
| Cbl | P22681 | Y700 | EGEEDTEYMTPSSRP | |
| Cbl | P22681 | Y731 | QQIDSCTYEAMYNIQ | |
| Cbl | P22681 | Y774 | SENEDDGYDVPKPPV | |
| DUSP3 (VHR) | P51452 | Y138 | SPTLVIAYLMMRQKM | + |
| DUSP3 (VHR) | P51452 | Y38 | NEVTPRIYVGNASVA | |
| FcGR2A | P12318 | Y281 | LEETNNDYETADGGY | |
| FcGR2A | P12318 | Y304 | TDDDKNIYLTLPPND | |
| FcGR2C | P31995 | Y287 | PEETNNDYETADGGY | |
| FcGR2C | P31995 | Y310 | TDDDKNIYLTLPPND | |
| GCET2 | Q8N6F7 | Y107 | GNSAEEYYENVPCKA | |
| GCET2 | Q8N6F7 | Y128 | LGGTETEYSLLHMPS | |
| HCA59 (C9orf78) | Q9NZ63 | Y147 | KNAEDCLYELPENIR | |
| HPK1 (MAP4K1) | Q92918 | Y381 | SESSDDDYDDVDIPT | + |
| HS1 | P14317 | Y378 | EPEPENDYEDVEEMD | |
| HS1 | P14317 | Y397 | EDEPEGDYEEVLEPE | + |
| ICLN (CLNS1A) | P54105 | Y214 | TEDSIRDYEDGMEVD | |
| IL15R alpha | Q13261 | Y227 | AVSLLACYLKSRQTP | |
| ITGB3 | P05106 | Y773 | DTANNPLYKEATSTF | |
| ITGB3 | P05106 | Y785 | STFTNITYRGI____ | |
| LAB | Q9GZY6 | Y136 | EDDDANSYENVLICK | |
| LAB | Q9GZY6 | Y193 | EDEESEDYQNSASIH | |
| LAB | Q9GZY6 | Y233 | EEDGEPDYVNGEVAA | |
| LAT | O43561 | Y200 | SMESIDDYVNVPESG | |
| LAT | O43561 | Y220 | SLDGSREYVNVSQEL | |
| LCP2 | Q13094 | Y113 | SSFEEDDYESPNDDQ | + |
| LCP2 | Q13094 | Y128 | DGEDDGDYESPNEEE | + |
| LCP2 | Q13094 | Y145 | PVEDDADYEPPPSND | + |
| MAPRE1 | Q15691 | Y124 | ANYDGKDYDPVAARQ | |
| NEK9 | Q8TD19 | Y520 | GLDSEEDYYTPQKVD | |
| NUCKS1 | Q9H1E3 | Y26 | SDDADEDYGRDSGPP | |
| PIK3AP1 (BCAP) | Q6ZUJ8 | Y195 | CGAETTVYVIVRCKL | |
| PIK3AP1 (BCAP) | Q6ZUJ8 | Y419 | GEEADAVYESMAHLS | |
| PIK3AP1 (BCAP) | Q6ZUJ8 | Y444 | PGCDEDLYESMAAFV | |
| PIK3AP1 (BCAP) | Q6ZUJ8 | Y459 | PAATEDLYVEMLQAS | |
| PKCa (PRKCA) | P17252 | Y657 | SDFEGFSYVNPQFVH | |
| PKCb (PRKCB) | P05771 | Y662 | NEFAGFSYTNPEFVI | |
| PLCG1 | P19174 | Y771 | IGTAEPDYGALYEGR | |
| PLCG1 | P19174 | Y783 | EGRNPGFYVEANPMP | + |
| PLCG2 | P16885 | Y759 | LYDVSRMYVDPSEIN | + |
| POU2AF1 (BOB1) | Q16633 | Y245 | EEEDSDAYALNHTLS | |
| RHBG | Q9H310-6 | Y429 | SPPDSQHYEDQVHWQ | |
| SH3BP2 (3BP2) | P78314 | S225 | SDMPRAHSFTSKGPG | |
| SH3BP2 (3BP2) | P78314 | S278 | PATPRRMSDPPLSTM | |
| SH3BP2 (3BP2) | P78314 | Y174 | YPTDNEDYEHDDEDD | |
| SH3BP2 (3BP2) | P78314 | Y183 | HDDEDDSYLEPDSPE | + |
| SH3BP2 (3BP2) | P78314 | Y448 | GDDSDEDYEKVPLPN | + |
| SLC4A1 | P02730 | Y21 | ENLEQEEYEDPDIPE | |
| SLC4A1 | P02730 | Y359 | AKPDSSFYKGLDLNG | |
| SLC4A1 | P02730 | Y8 | MEELQDDYEDMMEEN | |
| SLC4A1 | P02730 | Y904 | EEEGRDEYDEVAMPV | |
| SNCA | P37840 | Y125 | VDPDNEAYEMPSEEG | |
| SNCA | P37840 | Y133 | EMPSEEGYQDYEPEA | |
| SNCA | P37840 | Y136 | SEEGYQDYEPEA___ | |
| STAT5A | P42229 | Y964 | LAKAVDGYVKPQIKQ | |
| Syk | P43405 | Y131 | KENLIREYVKQTWNL | - |
| Syk | P43405 | Y323 | STVSFNPYEPELAPW | + |
| Syk | P43405 | Y348 | LPMDTEVYESPYADP | + |
| Syk | P43405 | Y352 | TEVYESPYADPEEIR | + |
| Syk | P43405 | Y525 | ALRADENYYKAQTHG | + |
| Syk | P43405 | Y526 | LRADENYYKAQTHGK | + |
| Tau | P10636 | Y17 | MEDHAGTYGLGDRKD | |
| TUBA1A | Q71U36 | Y432 | MAALEKDYEEVGVDS |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
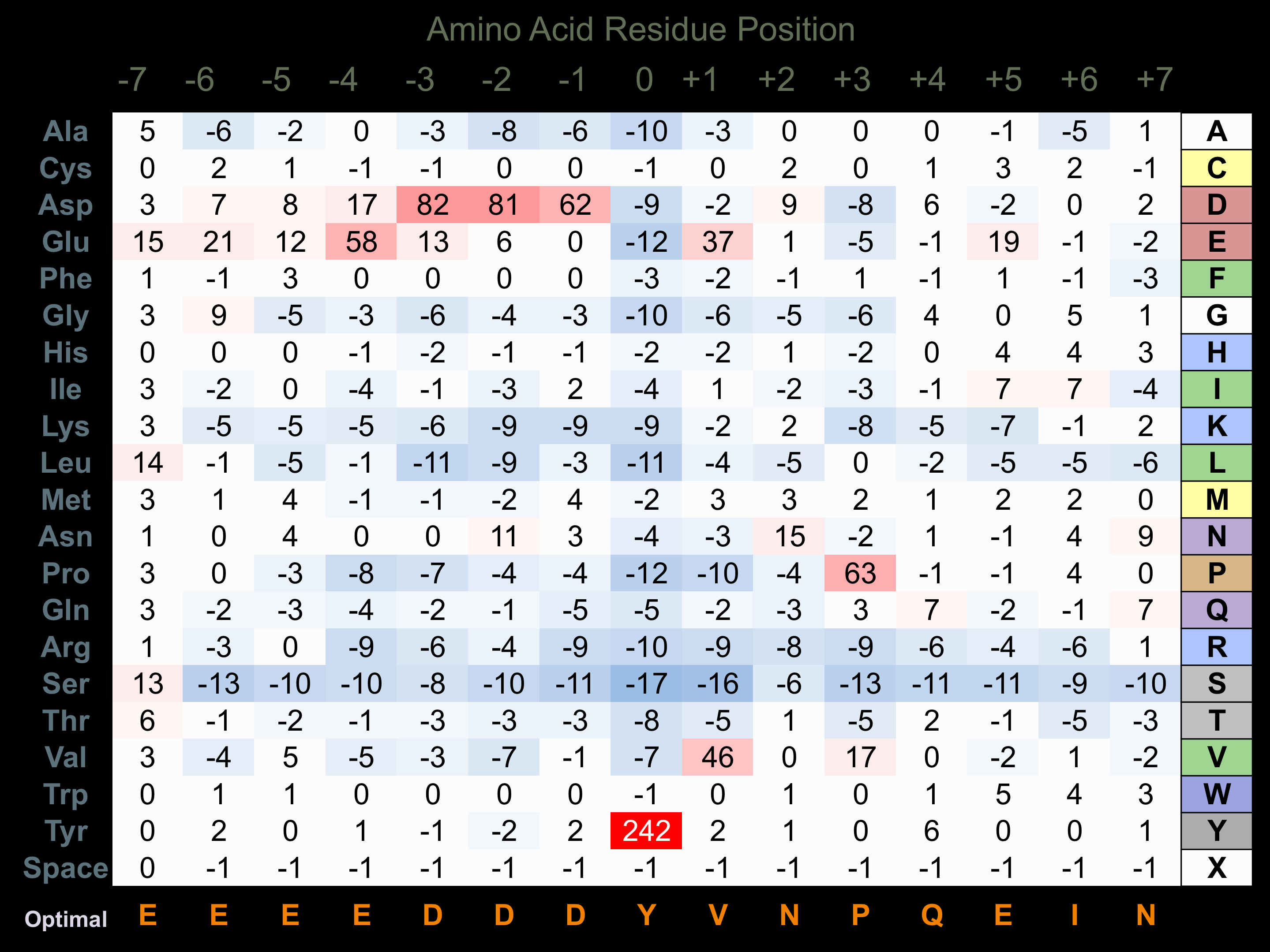
Matrix Type:
Experimentally derived from alignment of 64 known protein substrate phosphosites and 41 peptides phosphorylated by recombinant Syk in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Bone disorder and infectious disorder
Specific Diseases (Non-cancerous):
Cherubism (CRBM); Mycobacterium Abscessus
Comments:
Syk is a protein-tyrosine kinase that is widely expressed in hematopoietic cells, and is involved in coupling activated immunoreceptors that mediate diverse cellular responses.
Comments:
SYK appears to be an oncoprotein (OP) and a tumour suppressor protein (TSP). Syk is an oncoprotein in retinoblastoma, which is upregulated and is required for tumour cell survival. It is a potential tumour suppressor in human breast cancer. SYK is also an important regulator in acute myeloid leukemia.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Breast epithelial carcinomas (%CFC= -69, p<0.0009); Classical Hodgkin lymphomas (%CFC= -74, p<0.001); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +419, p<0.003); and Ovary adenocarcinomas (%CFC= +93, p<0.021) The COSMIC website notes an up-regulated expression score for SYK in diverse human cancers of 294, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 52 for this protein kinase in human cancers was 0.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25821 diverse cancer specimens. This rate is only 20 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.57 % in 805 skin cancers tested; 0.44 % in 1152 large intestine cancers tested; 0.29 % in 589 stomach cancers tested; 0.16 % in 1270 liver cancers tested; 0.13 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: A330T (8).
Comments:
Only 2 deletions, 4 insertions, and no complex mutations are noted on the COSMIC website.

