Nomenclature
Short Name:
PKCt
Full Name:
Protein kinase C, theta type
Alias:
- EC 2.7.11.13
- NPKC-theta
- PKCQ
- PKC-theta
- PRKCQ
- PRKCT; Protein kinase C, theta; Protein kinase C, theta type
- Kinase PKC-theta
- KPCT
- MGC126514
- MGC141919
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKC
SubFamily:
Delta
Specific Links
Structure
Mol. Mass (Da):
81,865
# Amino Acids:
706
# mRNA Isoforms:
3
mRNA Isoforms:
81,865 Da (706 AA; Q04759); 74,287 Da (643 AA; Q04759-2); 67,560 Da (581 AA; Q04759-3)
4D Structure:
Interacts with GLRX3 (via N-terminus)
1D Structure:
Subfamily Alignment
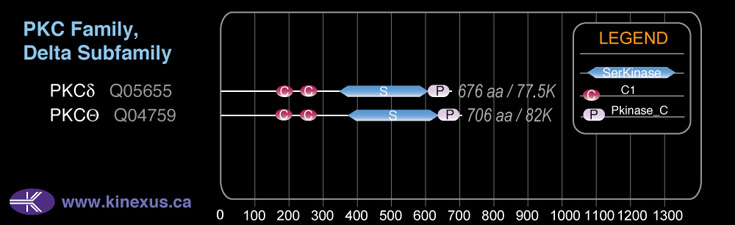
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K384.
Serine phosphorylated:
S323, S348, S370, S662, S676+, S685, S695+.
Threonine phosphorylated:
T51, T219, T275, T307, T536+, T538+, T542-.
Tyrosine phosphorylated:
Y28, Y90+, Y121, Y545-.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 58
58
710
32
975
 5
5
60
17
74
 3
3
32
18
25
 26
26
319
103
463
 31
31
381
24
302
 3
3
31
74
14
 21
21
253
35
428
 58
58
715
52
1571
 21
21
259
17
175
 10
10
120
104
85
 4
4
55
41
77
 51
51
633
193
713
 12
12
148
40
84
 5
5
59
12
53
 7
7
91
21
86
 3
3
41
15
40
 2
2
25
288
28
 5
5
66
26
59
 56
56
694
120
441
 22
22
271
109
311
 6
6
72
35
61
 8
8
93
38
67
 4
4
52
28
31
 5
5
59
27
88
 20
20
241
35
216
 53
53
648
65
729
 7
7
91
43
44
 4
4
53
28
54
 4
4
54
29
53
 23
23
286
28
155
 100
100
1233
24
795
 54
54
661
30
574
 4
4
50
69
204
 63
63
774
57
728
 12
12
154
35
100
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
99.7
100 33.6
33.6
45.1
99 -
-
-
96 -
-
-
- 96.7
96.7
98.3
97 -
-
-
- 94.7
94.7
97.5
95 94
94
96.8
94.5 -
-
-
- 86.1
86.1
92.9
- 63.3
63.3
77.1
83 33.9
33.9
48.2
- 34.1
34.1
49.2
75 -
-
-
- 42.9
42.9
58.7
- 43.2
43.2
58.9
- 47.1
47.1
64.1
- 47.8
47.8
62
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 27.2
27.2
39.3
- 26.5
26.5
40.2
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | VAV1 - P15498 |
| 2 | MARK3 - P27448 |
| 3 | IKBKB - O14920 |
| 4 | LCK - P06239 |
| 5 | BTK - Q06187 |
| 6 | AKT1 - P31749 |
| 7 | MSN - P26038 |
| 8 | CASP3 - P42574 |
| 9 | CARD11 - Q9BXL7 |
| 10 | RASGRP3 - Q8IV61 |
| 11 | PRKCZ - Q05513 |
| 12 | GLRX3 - O76003 |
| 13 | ICAM3 - P32942 |
| 14 | HABP4 - Q5JVS0 |
| 15 | BCL10 - O95999 |
Regulation
Activation:
Phosphorylation at Tyr-90, Thr-538 (activation loop of the kinase domain) and Ser-676 (turn motif) increases phosphotransferase activity. Phosphorylation at Ser-695 (hydrophobic region) increases phosphotransferase activity and interaction with PDK1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| CARD11 | Q9BXL7 | S559 | QPPRSRSSIMSITAE | |
| CARD11 | Q9BXL7 | S644 | NLMFRKFSLERPFRP | |
| CARD11 | Q9BXL7 | S645 | LERPFRPSVTSVGHV | |
| CD4 | P01730 | S433 | RRQAERMSQIKRLLS | |
| Ezrin | P15311 | T567 | QGRDKYKTLRQIRQG | |
| H3.1 | P68431 | S11 | TKQTARKSTGGKAPR | + |
| HePTP | P35236 | S134 | TILPNPQSRVCLGRA | |
| HePTP | P35236 | S246 | QYQEERRSVKHILFS | + |
| ICAM3 | P32942 | S518 | REHQRSGSYHVREES | |
| IRS1 | P35568 | S1101 | GCRRRHSSETFSSTP | - |
| IRS1 | P35568 | S323 | MVGGKPGSFRVRASS | |
| K-Ras iso2 | P01116-2 | S181 | GKKKKKKSKTKCVIM | |
| mGluR5 | P41594 | S840 | VRSAFTTSTVVRMHV | |
| Moesin | P26038 | T558 | LGRDKYKTLRQIRQG | |
| MYPT1 | O14974 | T853 | PREKRRSTGVSFWTQ | |
| NDRG2 | Q9UN36 | S332 | LSRSRTASLTSAASV | |
| PDK1 (PDPK1) | O15530 | S501 | LKGEIPWSQELRPEA | |
| PDK1 (PDPK1) | O15530 | S529 | TYYLMDPSGNAHKWC | |
| PKCt (PRKCQ) | Q04759 | S676 | LNEKPRLSFADRALI | + |
| PKCt (PRKCQ) | Q04759 | S695 | QNMFRNFSFMNPGME | + |
| PKCt (PRKCQ) | Q04759 | T219 | SAINSRETMFHKERF | ? |
| PTPN6 (SHP1) | P29350 | S591 | DKEKSKGSLKRK___ | |
| Radixin | P35241 | T564 | AGRDKYKTLRQIRQG | |
| RapGEF2 | Q9Y4G8 | S960 | KKRVRRSSFLNAKKL | |
| RasGRP1 | O95267 | T184 | RDWSRKLTQRIKSNT | |
| RasGRP3 | Q8IV61 | T133 | YDWMRRVTQRKKVSK | |
| STLK3 (STK39; SPAK) | Q9UEW8 | S311 | MMKKYGKSFRKLLSL | + |
| STLK3 (STK39; SPAK) | Q9UEW8 | S325 | LCLQKDPSKRPTAAE | + |
| WIP | O43516 | S488 | RNESRSGSNRRERGA |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
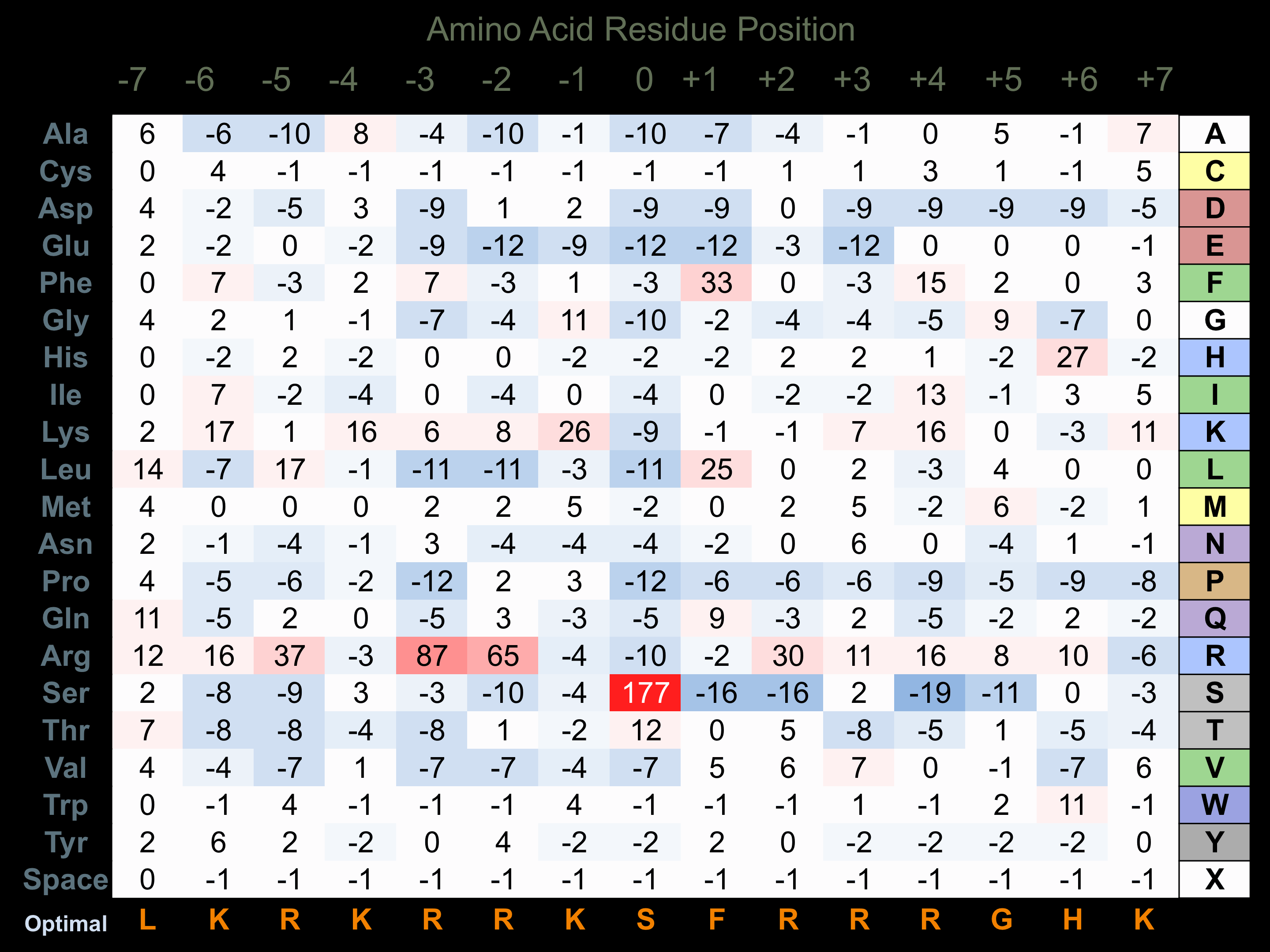
Matrix Type:
Experimentally derived from alignment of 45 known protein substrate phosphosites and 21 peptides phosphorylated by recombinant PKCt in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Gastrointestinal stromal tumour (GANT, GIST)
Comments:
PKCt is linked to Gastrointestinal Stromal Tumours (GANT, GIST), which are rare sarcomas arising from tissues lining the gastrointestinal tract and typically arise from the stomach, intestines, or colon. GANT has been related to neurofibromatosis and adenocarcinoma as well.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Colorectal adenocarcinomas (early onset) (%CFC= +83, p<0.01); Oral squamous cell carcinomas (OSCC) (%CFC= +47, p<0.004); Ovary adenocarcinomas (%CFC= +148, p<0.0009); and Skin fibrosarcomas (%CFC= +173, p<0.027). The COSMIC website notes an up-regulated expression score for PKCt in diverse human cancers of 349, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. The phosphotransferase activity of PKCt is unaffected by an Y90F mutation, but T-cell proliferation is inhibited. An A148E mutation can result in constitutive activation of PKCt. A T219A mutation also does not inhibit kinase phosphotransferase activity, but it will inhibit localization to the plasma membrane and inhibit transactivation of IL2 promoter. A complete loss of phosphotramsferase activity occurs with either a K409A/E or T538A mutation, while a S676A or S695A mutation can lead to reduced kinase activity.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 25349 diverse cancer specimens. This rate is a modest 1.59-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.84 % in 805 skin cancers tested; 0.49 % in 1094 large intestine cancers tested; 0.38 % in 602 endometrium cancers tested; 0.36 % in 589 stomach cancers tested; 0.16 % in 904 ovary cancers tested; 0.14 % in 1941 lung cancers tested; 0.13 % in 1466 breast cancers tested; 0.12 % in 1270 liver cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: I46M (6).
Comments:
Only 1 deletion, 3 insertions and no complex mutations are noted on the COSMIC website. About 27% of the point mutations are silent and do not change the amino acid sequence of the protein kinase.

