Nomenclature
Short Name:
MEK5
Full Name:
Dual specificity mitogen-activated protein kinase kinase 5
Alias:
- CXA1
- EC 2.7.12.2
- MAPK/ERK kinase 5
- MAPKK5
- MKK5
- PRKMK5
- HsT17454
- MAP kinase kinase 5
- MAP2K5
- MAPK,ERK kinase 5
Classification
Type:
Dual specificity protein kinase
Group:
STE
Family:
STE7
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
48,968
# Amino Acids:
438
# mRNA Isoforms:
4
mRNA Isoforms:
50,112 Da (448 AA; Q13163); 49,497 Da (444 AA; Q13163-3); 48,968 Da (438 AA; Q13163-2); 46,131 Da (412 AA; Q13163-4)
4D Structure:
Interacts with PARD6A, MAP3K3 and MAPK7. Forms a complex with SQSTM1 and PRKCZ or PRKCI By similarity. Interacts with Yersinia yopJ.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
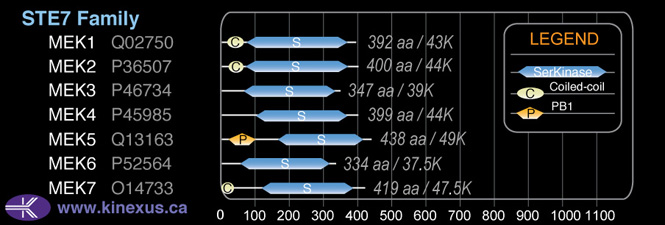
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Threonine phosphorylated:
T71+, T315+, .
Tyrosine phosphorylated:
Y316+.
Serine phosphorylated:
S129, S133, S137, S142, S148, S149, S311+.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 96
96
638
81
876
 7
7
47
40
41
 14
14
95
26
102
 41
41
273
255
445
 87
87
575
66
494
 4
4
24
227
41
 39
39
261
95
461
 73
73
486
103
548
 57
57
378
45
277
 11
11
74
257
109
 8
8
50
81
56
 77
77
511
524
552
 6
6
43
92
56
 6
6
43
34
39
 10
10
67
65
72
 7
7
44
46
48
 8
8
51
511
279
 14
14
91
47
93
 9
9
61
264
78
 61
61
402
321
464
 13
13
86
60
78
 11
11
70
67
78
 11
11
76
56
75
 13
13
85
50
91
 19
19
128
58
160
 73
73
483
149
557
 6
6
41
95
56
 10
10
63
47
50
 11
11
72
50
90
 9
9
57
84
50
 60
60
394
48
649
 93
93
614
102
1092
 24
24
157
162
478
 100
100
662
171
634
 32
32
213
78
241
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 93.4
93.4
93.4
100 99.5
99.5
99.5
100 -
-
-
99 -
-
-
94 63.4
63.4
64.3
99 -
-
-
- 97.8
97.8
99.1
98 98.2
98.2
99.1
98 -
-
-
- 82.4
82.4
86.2
- 74.1
74.1
76.1
94 84.8
84.8
90
85 30.6
30.6
47.3
79 -
-
-
- 35
35
54.2
- -
-
-
- 37
37
53.8
- 49.3
49.3
63.2
- -
-
-
- -
-
-
- -
-
-
- 30.8
30.8
47.5
- 27.5
27.5
42.7
40 -
-
-
40
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BAD - Q92934 |
| 2 | PRKCI - P41743 |
| 3 | MAPK7 - Q13164 |
| 4 | GSK3B - P49841 |
| 5 | PARD6A - Q9NPB6 |
| 6 | MAPK1 - P28482 |
Regulation
Activation:
Activated by phosphorylation on Ser/Thr by MAP kinase kinase kinases. ; Activation of this pathway appear to play a critical role in protecting cells from stress-induced apopotosis, neuronal survival and cardiac development and angiogenesis.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ERK5 (MAPK7) | Q13164 | T219 | AEHQYFMTEYVATRW | + |
| ERK5 (MAPK7) | Q13164 | Y221 | HQYFMTEYVATRWYR | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
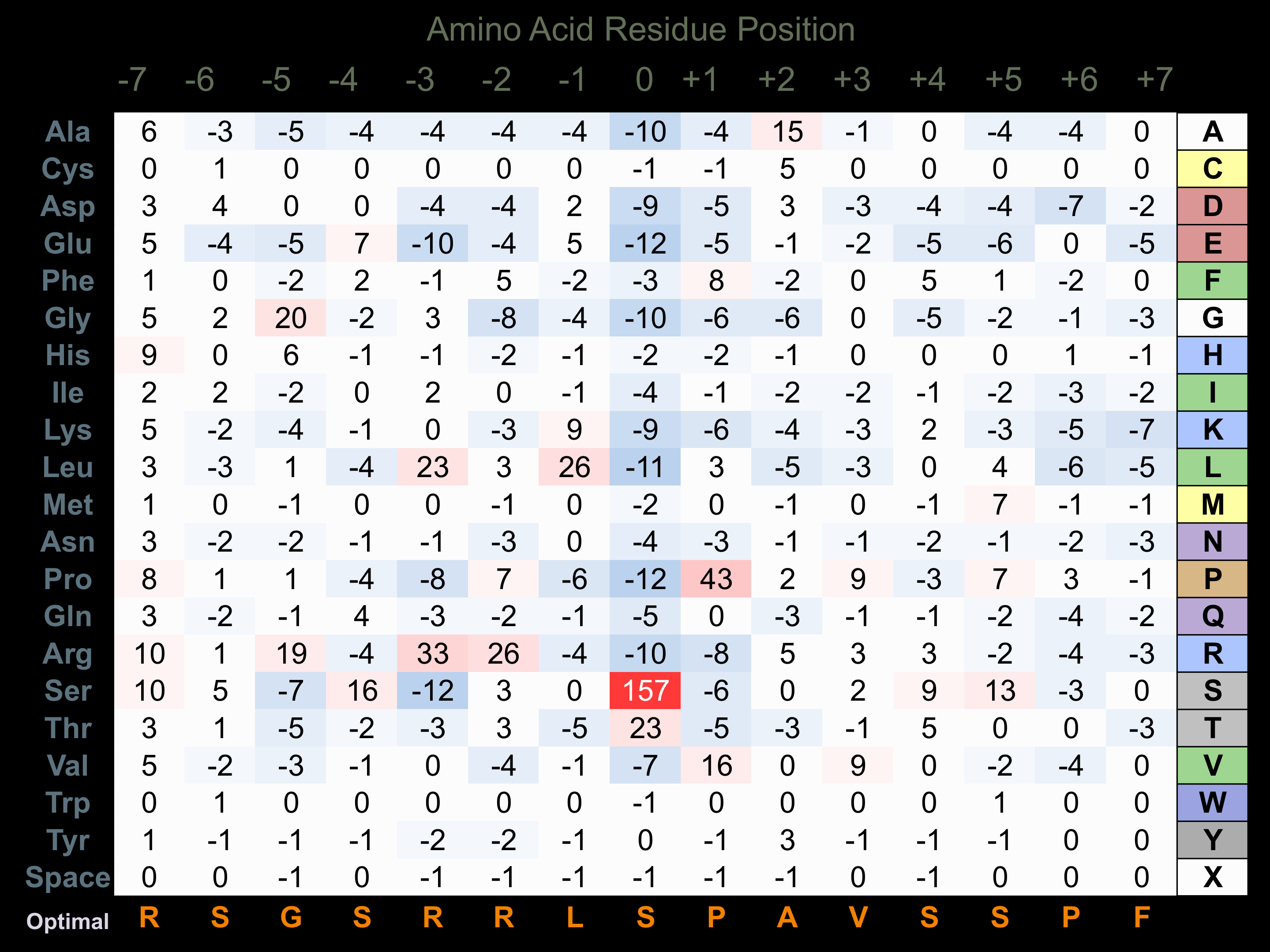
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Developmental, and motor control disorders
Specific Diseases (Non-cancerous):
Developmental coordination disorder (DCD)
Comments:
Developmental Coordination Disorder (DCD) is characterized by uncoordinated muscle movement. Affected tissues with DCD can include the brain, testes, and eye. MEK5 has been inactivated with the K195M, or S311A, or T315A mutations.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer stage 2A (%CFC= +85, p<0.066); Cervical cancer stage 2B (%CFC= +102, p<0.072); Skin melanomas (%CFC= -54, p<0.043); and Uterine leiomyomas (%CFC= +109, p<0.039).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24454 diverse cancer specimens. This rate is only -12 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.33 % in 1052 large intestine cancers tested; 0.14 % in 1594 lung cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,223 cancer specimens
Comments:
Only 2 deletions and 1 insertion, and no complex mutations are noted on the COSMIC website.

