Nomenclature
Short Name:
ERK5
Full Name:
Mitogen-activated protein kinase 7
Alias:
- BMK
- Extracellular-signal-regulated kinase 5
- Kinase ERK5
- MAPK7
- PRKM7
- BMK1
- BMK1 kinase
- EC 2.7.11.24
- ERK4
- ERK-5
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
MAPK
SubFamily:
ERK
Specific Links
Structure
Mol. Mass (Da):
88386
# Amino Acids:
816
# mRNA Isoforms:
4
mRNA Isoforms:
88,386 Da (816 AA; Q13164); 73,218 Da (677 AA; Q13164-2); 59,328 Da (533 AA; Q13164-3); 50,152 Da (451 AA; Q13164-4)
4D Structure:
Interacts with MAP2K5. Forms oligomers By similarity. Interacts with MEF2A, MEF2C and MEF2D; the interaction phosphorylates the MEF2s and enhances transcriptional activity of MEF2A, MEF2C but not MEF2D
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
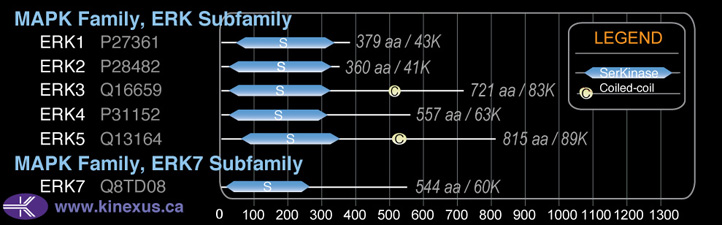
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 55 | 347 | Pkinase |
| 507 | 543 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
A2, K35.
Serine phosphorylated:
S31, S70, S71, S282, S322, S420, S421, S432, S433, S472, S485, S486-, S495, S496, S562, S567, S707, S718, S720, S730, S731, S754, S769, S770, S772, S773, S774, S775, S776, S803.
Threonine phosphorylated:
T28, T219+, T224, T280, T583, T584, T732, T733.
Tyrosine phosphorylated:
Y216+, Y221+.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 78
78
740
32
1019
 6
6
61
15
38
 40
40
378
8
923
 67
67
632
87
1661
 69
69
652
25
570
 4
4
38
80
42
 24
24
232
31
521
 53
53
503
37
630
 31
31
292
17
232
 11
11
107
79
315
 9
9
87
27
264
 74
74
704
169
711
 6
6
58
30
155
 5
5
51
12
50
 12
12
110
21
358
 9
9
83
15
48
 29
29
273
116
2281
 18
18
169
17
548
 5
5
52
83
180
 54
54
510
109
537
 9
9
90
19
194
 17
17
158
23
377
 7
7
71
18
138
 15
15
147
17
316
 25
25
238
19
806
 100
100
950
53
1536
 10
10
93
33
369
 10
10
92
17
230
 10
10
92
17
187
 9
9
84
28
36
 68
68
642
36
522
 59
59
563
36
577
 47
47
445
73
606
 80
80
756
57
675
 6
6
60
44
58
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 54.7
54.7
54.7
99 97.7
97.7
98
97.5 -
-
-
98 -
-
-
91 86.8
86.8
88
98 -
-
-
- 92
92
93.3
92 92.2
92.2
93.4
91 -
-
-
- 30.4
30.4
31
- -
-
-
- 24.1
24.1
32.8
72 47.3
47.3
59.2
69 -
-
-
- -
-
-
- -
-
-
- -
-
-
47 32.7
32.7
46.7
- 26.2
26.2
35
- 26.1
26.1
35
- -
-
-
54 25.7
25.7
34.7
53 28.8
28.8
40.6
49 -
-
-
52
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | GJA1 - P17302 |
| 2 | MEF2D - Q14814 |
| 3 | MEF2A - Q02078 |
| 4 | SGK1 - O00141 |
| 5 | MYC - P01106 |
| 6 | YWHAE - P62258 |
| 7 | RXRA - P19793 |
| 8 | YWHAB - P31946 |
| 9 | PTPRR - Q15256 |
| 10 | RAF1 - P04049 |
| 11 | ETS1 - P14921 |
| 12 | MAP2K5 - Q13163 |
| 13 | PPP2R4 - Q15257 |
Regulation
Activation:
Phosphorylation of Ser-311 and Thr-315 increases phosphotransferase activity and inhibits interaction with ERK5. Activated in response to hyperosmolarity, hydrogen peroxide, and epidermal growth factor (EGF).
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Cx43 | P17302 | S254 | HATSGALSPAKDCGS | |
| ERK5 (MAPK7) | Q13164 | S421 | CPDVEMPSPWAPSGD | |
| ERK5 (MAPK7) | Q13164 | S433 | SGDCAMESPPPAPPP | |
| ERK5 (MAPK7) | Q13164 | S496 | SRLRDGPSAPLEAPE | |
| ERK5 (MAPK7) | Q13164 | S731 | DPLPPVFSGTPKGSG | |
| ERK5 (MAPK7) | Q13164 | T28 | VKAEPAHTAASVAAK | |
| ERK5 (MAPK7) | Q13164 | T733 | LPPVFSGTPKGSGAG | |
| Ets-1 | P14921 | T38 | CADVPLLTPSSKEMM | + |
| Fos | P01100 | S32 | DSLSYYHSPADSFSS | |
| Fos | P01100 | T232 | GGLPEVATPESEEAF | + |
| MAPKAPK5 | Q8IW41 | T182 | IDQGDLMTPQFTPYY | + |
| MEF2A | Q02078 | S355 | SALQGFNSPGMLSLG | + |
| MEF2A | Q02078 | T312 | QATQPLATPVVSVTT | + |
| MEF2A | Q02078 | T319 | TPVVSVTTPSLPPQG | + |
| MEF2C iso3 | Q06413 | S387 | TRHEAGRSPVDSLSS | + |
| MEF2D | Q14814 | S180 | LTDPRLLSPQQPALQ | + |
| MEK5 (MAP2K5, MKK5) | Q13163 | S129 | VNTRAGPSQHSSPAV | |
| MEK5 (MAP2K5, MKK5) | Q13163 | S137 | QHSSPAVSDSLPSNS | |
| MEK5 (MAP2K5, MKK5) | Q13163 | S142 | AVSDSLPSNSLKKSS | |
| MEK5 (MAP2K5, MKK5) | Q13163 | S149 | SNSLKKSSAELKKIL | |
| Neurogenin 1 | Q92886 | S178 | PCLPGPPSPASDAES | |
| Neurogenin 1 | Q92886 | S201 | SPLSDPSSPAASEDF | |
| NFAT3 | Q14934 | S168 | QGGGAFFSPSPGSSS | - |
| NFAT3 | Q14934 | S170 | GGAFFSPSPGSSSLS | - |
| SGK1 | O00141 | S78 | ANPSPPPSPSQQINL | ? |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
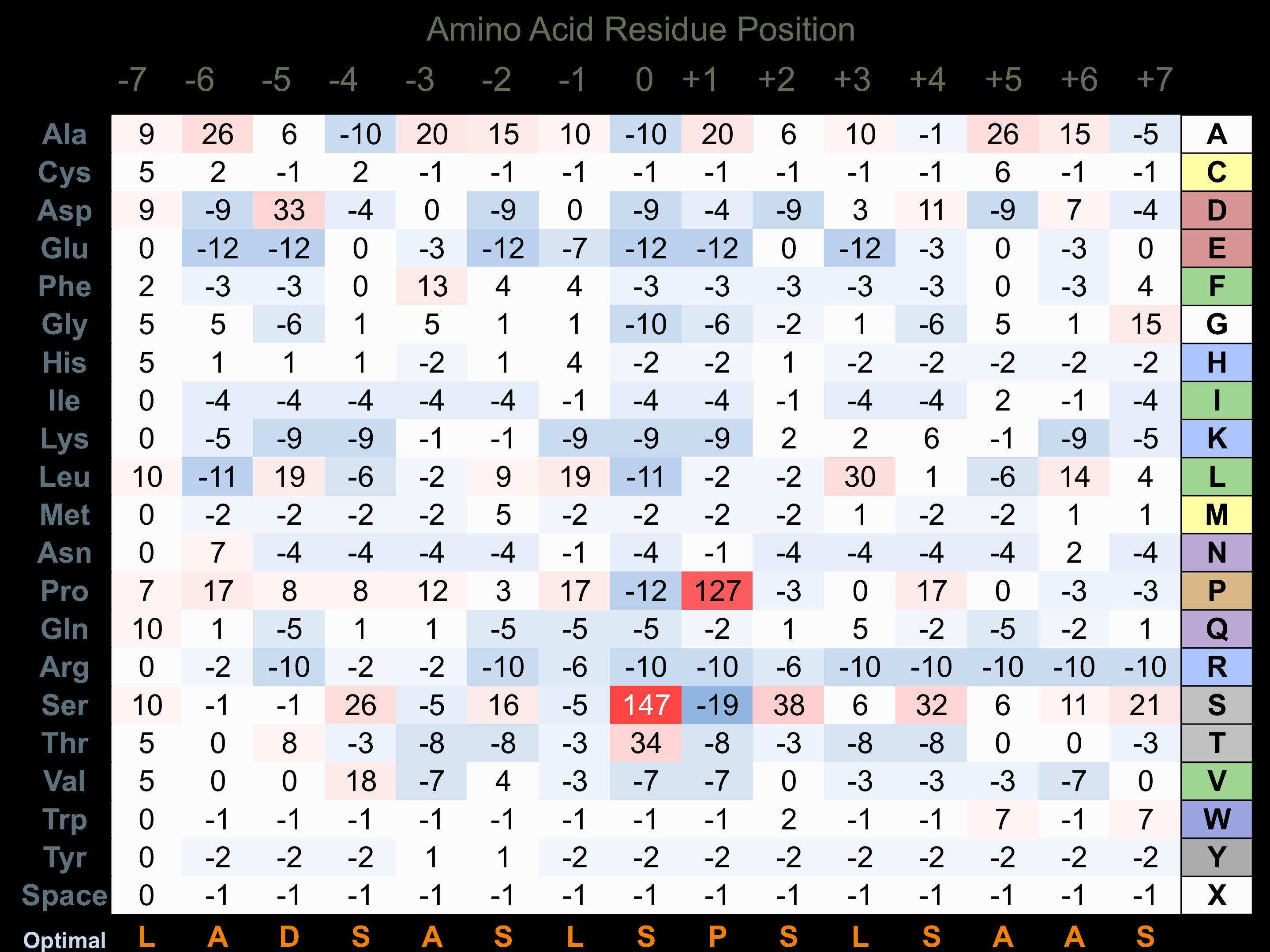
Matrix Type:
Experimentally derived from alignment of 37 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AT7519 | Kd = 42 nM | 11338033 | 22037378 | |
| NVP-TAE684 | Kd = 48 nM | 16038120 | 509032 | 22037378 |
| Aurora A Inhibitor 1 (DF) | Kd < 50 nM | 21992004 | ||
| Aurora A Inhibitor 23 (DF) | Kd < 50 nM | 21992004 | ||
| BIX02188 | IC50 = 59 nM | 23507698 | 18834865 | |
| TTT-3002 | IC50 < 60 nM | |||
| TG101348 | Kd = 66 nM | 16722836 | 1287853 | 22037378 |
| XMD8-92 | IC50 = 80 nM | 46843772 | 1673046 | 20832753 |
| R547 | Kd = 110 nM | 6918852 | 22037378 | |
| Aurora A Inhibitor 29 (DF) | Kd < 400 nM | 21992004 | ||
| Crizotinib | Kd = 510 nM | 11626560 | 601719 | 22037378 |
| JNJ-7706621 | Kd = 520 nM | 5330790 | 191003 | 18183025 |
| SNS032 | Kd = 650 nM | 3025986 | 296468 | 18183025 |
| BIX02188 | IC50 = 830 nM | 23507698 | 18834865 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Lestaurtinib | Kd = 1.1 µM | 126565 | 22037378 | |
| Staurosporine | Kd = 1.6 µM | 5279 | 18183025 | |
| PHA-665752 | Kd = 1.7 µM | 10461815 | 450786 | 22037378 |
| Bosutinib | Kd = 2.3 µM | 5328940 | 288441 | 22037378 |
| Nintedanib | Kd = 2.5 µM | 9809715 | 502835 | 22037378 |
| KW2449 | Kd = 2.7 µM | 11427553 | 1908397 | 22037378 |
| BI2536 | Kd = 3.2 µM | 11364421 | 513909 | 22037378 |
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Osteosarcomas
Comments:
ERK5 may be a tumour requiring protein (TRP). Although there has been no direct linkage of ERK5 to formation of carcinomas in humans in vivo, in vitro ERK5 over-expression has been shown to increase metastasis, and ERK5 inhibition has been shown to inhibit osteosarcoma cell motility.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -76, p<0.006); Brain oligodendrogliomas (%CFC= -60, p<0.009); Breast epithelial carcinomas (%CFC= -80, p<0.017); Cervical cancer stage 2A (%CFC= +79, p<0.097); and Oral squamous cell carcinomas (OSCC) (%CFC= -56, p<0.074). The COSMIC website notes an up-regulated expression score for ERK5 in diverse human cancers of 378, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 184 for this protein kinase in human cancers was 3.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. Activation the phosphotransferase activity of ERK5, through MAP2K5, can be inhibited with the mutations T219A+Y221F.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25371 diverse cancer specimens. This rate is only -19 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.3 % in 805 skin cancers tested; 0.29 % in 1093 large intestine cancers tested; 0.26 % in 602 endometrium cancers tested; 0.21 % in 589 stomach cancers tested; 0.09 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,654 cancer specimens
Comments:
Only 2 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.

