Nomenclature
Short Name:
DYRK1A
Full Name:
Dual-specificity tyrosine-phosphorylation regulated kinase 1A
Alias:
- DYR1A
- DYRK
- EC 2.7.12.1
- EC 2.7.12.2
- MNB
- MNBH
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
DYRK
SubFamily:
Dyrk1
Specific Links
Structure
Mol. Mass (Da):
85,584
# Amino Acids:
763
# mRNA Isoforms:
5
mRNA Isoforms:
85,584 Da (763 AA; Q13627); 84,556 Da (754 AA; Q13627-2); 66,099 Da (584 AA; Q13627-5); 61,770 Da (540 AA; Q13627-4); 60,330 Da (529 AA; Q13627-3)
4D Structure:
Interacts RAD54L2/ARIP4 By similarity. Interacts with RANBP9. Interacts with WDR68
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 159 | 479 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K105.
Serine phosphorylated:
S14, S22, S127, S324, S529+, S538, S555, S706, S748, S758, S762.
Tyrosine phosphorylated:
Y111, Y112, Y136, Y145, Y147, Y159, Y219, Y319, Y321+, Y705.
Ubiquitinated:
K453.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1542
45
2490
 5
5
84
25
85
 12
12
184
3
277
 28
28
433
129
539
 45
45
699
36
641
 10
10
153
118
213
 30
30
470
55
580
 42
42
650
53
918
 34
34
530
24
548
 8
8
121
140
137
 6
6
85
36
106
 38
38
585
309
634
 9
9
144
36
195
 10
10
157
20
171
 7
7
101
28
104
 9
9
132
23
157
 7
7
115
379
133
 6
6
96
18
76
 16
16
253
136
346
 35
35
540
162
667
 11
11
165
24
182
 13
13
205
30
306
 4
4
63
30
90
 11
11
162
20
158
 18
18
270
27
400
 52
52
802
72
909
 8
8
117
39
147
 7
7
113
18
110
 12
12
183
16
190
 13
13
200
42
139
 47
47
728
54
721
 83
83
1276
51
2140
 14
14
216
86
326
 51
51
782
93
725
 21
21
328
66
573
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 99.6
99.6
99.8
100 -
-
-
100 -
-
-
98 85.1
85.1
85.5
99.5 -
-
-
- 99.2
99.2
99.4
99 99.6
99.6
99.7
100 -
-
-
- 94.4
94.4
95.2
- 26.7
26.7
39.9
96 91.4
91.4
94.2
93.5 78.7
78.7
84.2
83 -
-
-
- 47.7
47.7
60.2
79 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | DCAF7 - P61962 |
| 2 | GLI1 - P08151 |
| 3 | PHYHIP - Q92561 |
| 4 | YWHAE - P62258 |
| 5 | CREB1 - P16220 |
| 6 | SFRS16 - Q8N2M8 |
| 7 | YWHAG - P61981 |
Regulation
Activation:
Phosphorylation at Tyr-321 increases phosphotransferase activity. Phosphorylation at Ser-529 increases phosphotransferase activity and association with 14-3-3-beta.
Inhibition:
Inhibited by RANBP9.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
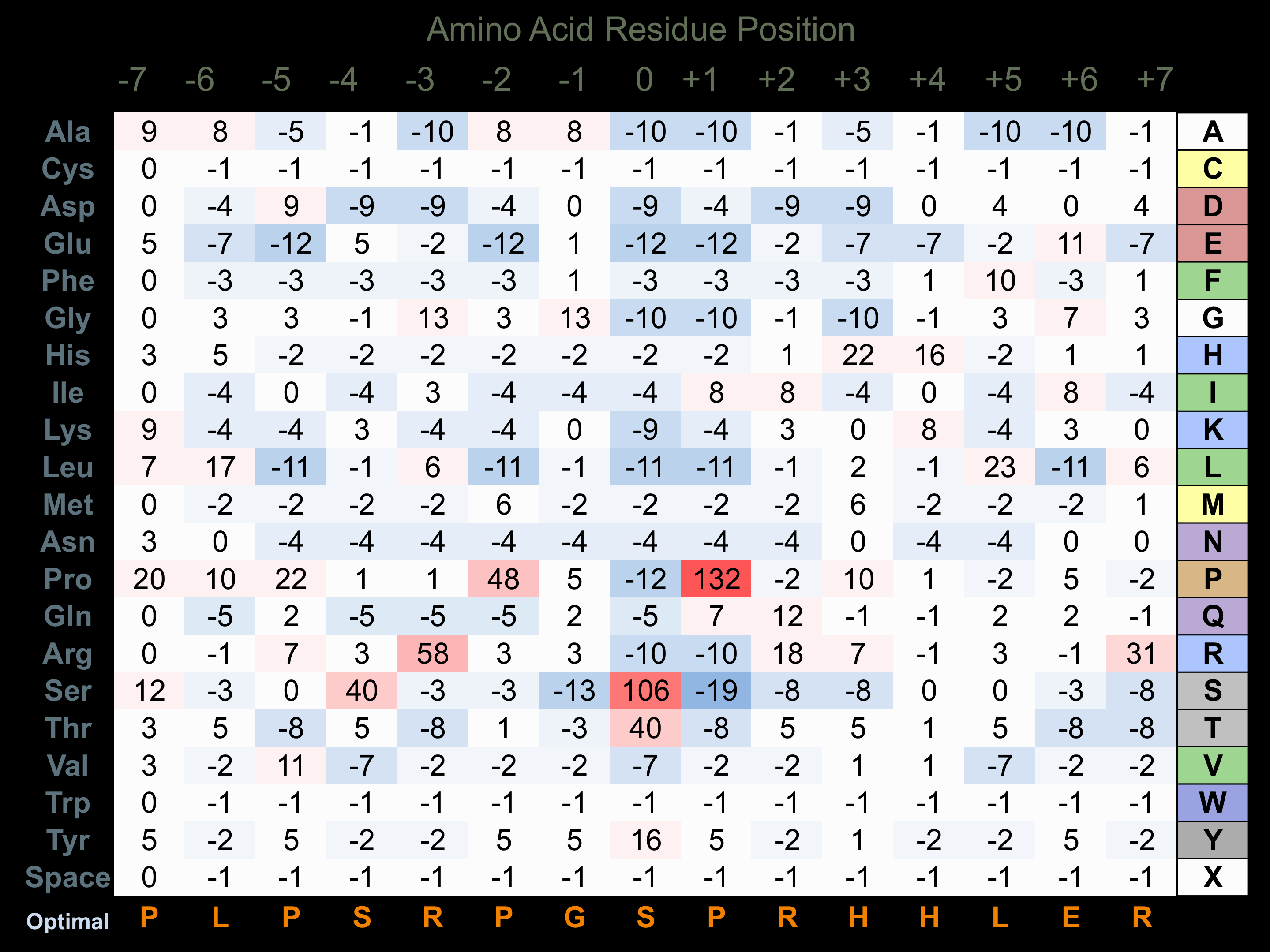
Matrix Type:
Experimentally derived from alignment of 33 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, cognitive impairment
Specific Diseases (Non-cancerous):
Down syndrome; Mental retardation, autosomal dominant 7 (MRD7); Mental retardation, autosomal dominant 10
Comments:
Down Syndrome is a rare chromosome condition leading to cognitive impairment, distinct facial characteristics, and other health defects. Down Syndrome may have a relation to Alzheimer's disease as well. The phenotype of Mental retardation, autosomal dominant 7 (MRD7) disorder includes microcephaly, severe cognitive impairment (lacking speech), anxious autistic behaviour, and distinct facial characteristics. Kinase phosphotransferase activity of DYRK1A can be abrogated with the K188R mutation, or it can be mildly impaired with a Y321F mutation (which still maintains autophosphorylation of tyrosine residues).
Specific Cancer Types:
Megakaryocytic tumour, Epithelial and Lymphoid tumours
Comments:
Megakaryocytic tumours (derived from bone marrow cells responsible for producing red blood cells) can be induced with DYRK1A.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for DYRK1A in diverse human cancers of 408, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 348 for this protein kinase in human cancers was 5.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25183 diverse cancer specimens. This rate is very similar (+ 7% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.41 % in 1093 large intestine cancers tested; 0.31 % in 589 stomach cancers tested; 0.3 % in 602 endometrium cancers tested; 0.13 % in 1753 lung cancers tested; 0.09 % in 1466 breast cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,466 cancer specimens
Comments:
Only 3 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.

