Nomenclature
Short Name:
BRK
Full Name:
Tyrosine-protein kinase 6
Alias:
- Brk6
- EC 2.7.10.2
- PTK6
- Tyrosine-protein kinase 6
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Src
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
51,834
# Amino Acids:
451
# mRNA Isoforms:
2
mRNA Isoforms:
51,834 Da (451 AA; Q13882); 14,465 Da (134 AA; Q13882-2)
4D Structure:
Interacts with GAP-A.p65. Interacts with KHDRBS1. Interacts with phosphorylated IRS4. Interacts with ADAM15.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
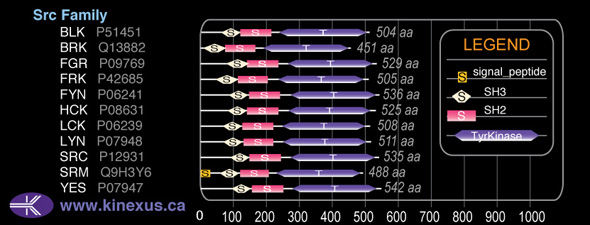
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S111, S117, S159, S279, S344, S446.
Threonine phosphorylated:
T445.
Tyrosine phosphorylated:
Y114, Y127, Y342+, Y351-, Y447- .
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1013
22
1508
 1.3
1.3
13
11
13
 22
22
221
8
582
 37
37
370
77
1231
 70
70
709
19
433
 8
8
79
50
77
 47
47
475
33
560
 28
28
282
26
419
 51
51
521
10
488
 9
9
92
61
136
 4
4
38
23
100
 40
40
402
118
512
 4
4
38
19
136
 0.9
0.9
9
9
6
 3
3
33
13
24
 1.4
1.4
14
13
17
 2
2
25
193
61
 7
7
69
15
113
 2
2
18
63
45
 90
90
907
79
725
 4
4
40
19
104
 4
4
43
19
139
 27
27
271
17
230
 3
3
30
15
86
 4
4
45
19
110
 98
98
997
49
2201
 8
8
77
22
152
 8
8
78
15
150
 13
13
132
14
446
 2
2
19
14
18
 31
31
313
18
228
 30
30
300
26
329
 10
10
102
61
223
 82
82
826
57
761
 7
7
73
35
117
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
98 78.7
78.7
80.9
- -
-
-
81.5 -
-
-
- 39.7
39.7
52.9
- -
-
-
- 80
80
89.8
80 39.1
39.1
52.7
80 -
-
-
- 67.9
67.9
80.5
- 37.8
37.8
53.9
- 38.7
38.7
53.7
52 42.4
42.4
60.4
49 -
-
-
- 40.8
40.8
55.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | ERBB3 - P21860 |
| 2 | EGFR - P00533 |
| 3 | STAP2 - Q9UGK3 |
| 4 | ERBB2 - P04626 |
Regulation
Activation:
Phosphorylation at Tyr-342 increases phosphotransferase activity. Phosphorylation at Tyr-447 increases phosphotransferase activity and inhibits interaction with PKBa/Akt1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Akt1 (PKBa) | P31749 | Y326 | EVLEDNDYGRAVDWW | + |
| ARHGAP5 | Q13017 | Y1109 | KGYSDEIYVVPDDSQ | |
| Brk (PTK6) | Q13882 | S159 | VNYHRAQSLSHGLRL | |
| Brk (PTK6) | Q13882 | S446 | ERLSSFTSYENPT__ | |
| Brk (PTK6) | Q13882 | Y114 | SEKPSADYVLSVRDT | |
| Brk (PTK6) | Q13882 | Y342 | RLIKEDVYLSHDHNI | + |
| Brk (PTK6) | Q13882 | Y351 | SHDHNIPYKWTAPEA | - |
| Brk (PTK6) | Q13882 | Y447 | RLSSFTSYENPT___ | - |
| Paxillin (PXN) | P49023 | Y118 | VGEEEHVYSFPNKQK | + |
| Paxillin (PXN) | P49023 | Y31 | FLSEETPYSYPTGNH | |
| Sam68 | Q07666 | Y435 | ARPVKGAYREHPYGR | |
| Sam68 | Q07666 | Y440 | GAYREHPYGRY____ | |
| Sam68 | Q07666 | Y443 | REHPYGRY_______ | |
| STAP2 | Q9UGK3 | Y250 | PFLLDEDYEKVLGYV | ? |
| STAT5B | P51692 | Y699 | TAKAVDGYVKPQIKQ | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
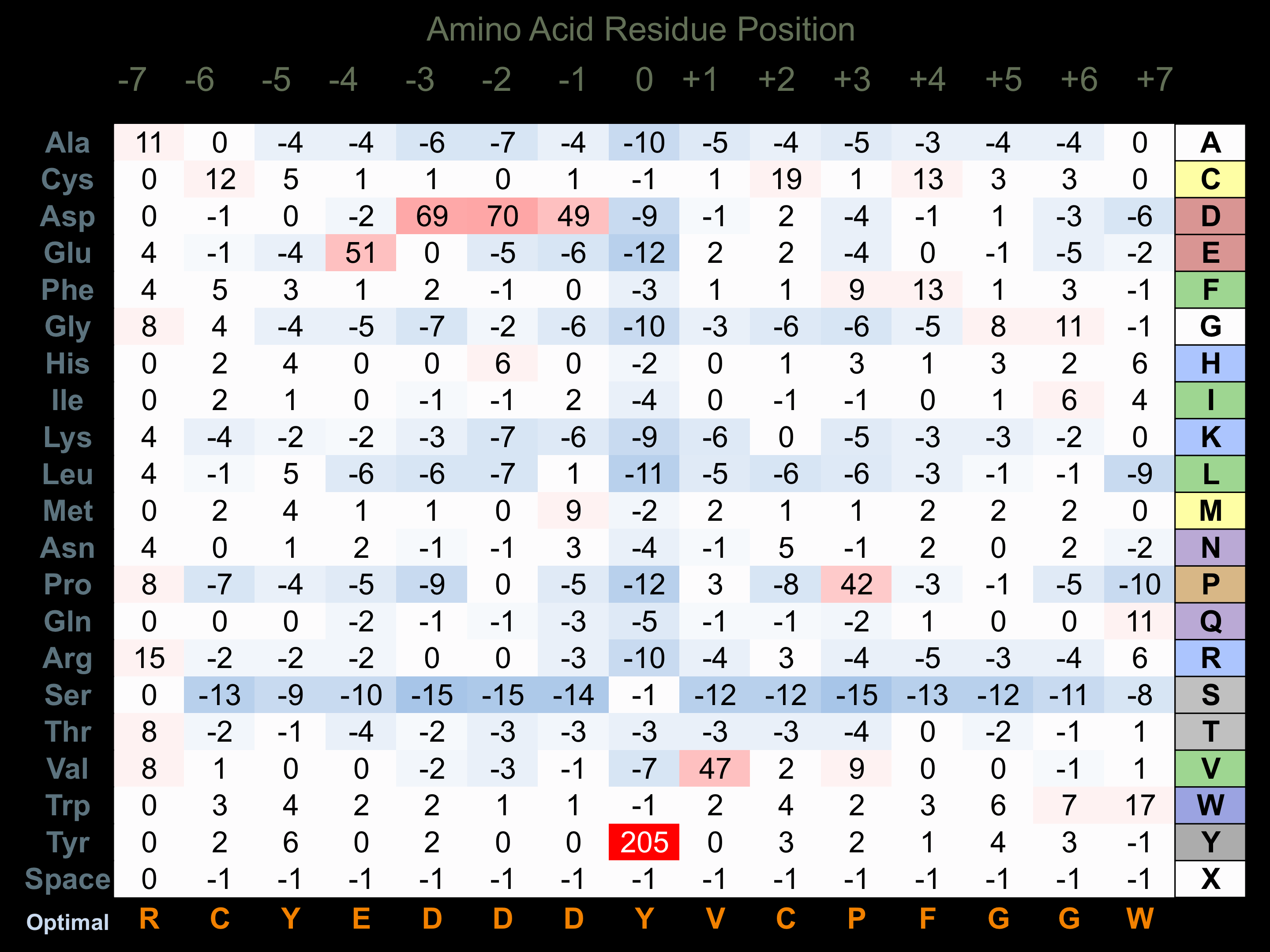
Matrix Type:
Experimentally derived from alignment of 20 known protein substrate phosphosites and 96 peptides phosphorylated by recombinant Brk in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Eye disorders
Specific Diseases (Non-cancerous):
Recurrent corneal erosion
Comments:
Recurrent corneal erosion is an eye disease characterized by failure of the outermost epithelial cell layer in the cornea to firmly adhere to the underlying membrane (Bownman's layer). The disease is very painful as the erosion of these epithelial cells results in the exposure of the underlying corneal nerves, which are very sensitive to even mild stimulation.
Comments:
Low levels of BRK expression have been reported in breast cancer tissues, which is not observed in normal breast tissue. Overexpression of BRK in mammalian epithelial cells leads to sensitization to EGF signalling, leading to the partially transformed phenotype. In addition, BRK has been shown to coimmunoprecipitate with the epidermal growth factor receptor, indicating direct interaction.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -58, p<0.011); Bladder carcinomas (%CFC= +202, p<0.005); Cervical epithelial cancer (%CFC= +86, p<0.005); Cervical cancer (%CFC= +335, p<0.0002); Colorectal adenocarcinomas (early onset) (%CFC= -46, p<0.0004); Oral squamous cell carcinomas (OSCC) (%CFC= -76, p<0.03); Skin melanomas (%CFC= -51, p<0.0004); and Skin melanomas - malignant (%CFC= -88, p<0.0001). The COSMIC website notes an up-regulated expression score for BRK in diverse human cancers of 589, which is 1.3-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 3 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25065 diverse cancer specimens. This rate is only -17 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.36 % in 854 skin cancers tested; 0.24 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,228 cancer specimens
Comments:
Only 2 deletion, 1 complex and no insertions mutations are noted on the COSMIC website.

