Nomenclature
Short Name:
ErbB4
Full Name:
Receptor tyrosine-protein kinase erbB-4
Alias:
- EC 2.7.10.1
- HER4
- Kinase ErbB4
- P180erbB4
- P180-erbB4
- V-erb-a erythroblastic leukemia viral oncogene 4
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
EGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
146,808
# Amino Acids:
1308
# mRNA Isoforms:
4
mRNA Isoforms:
146,808 Da (1308 AA; Q15303); 145,578 Da (1298 AA; Q15303-2); 145,198 Da (1292 AA; Q15303-3); 143,968 Da (1282 AA; Q15303-4)
4D Structure:
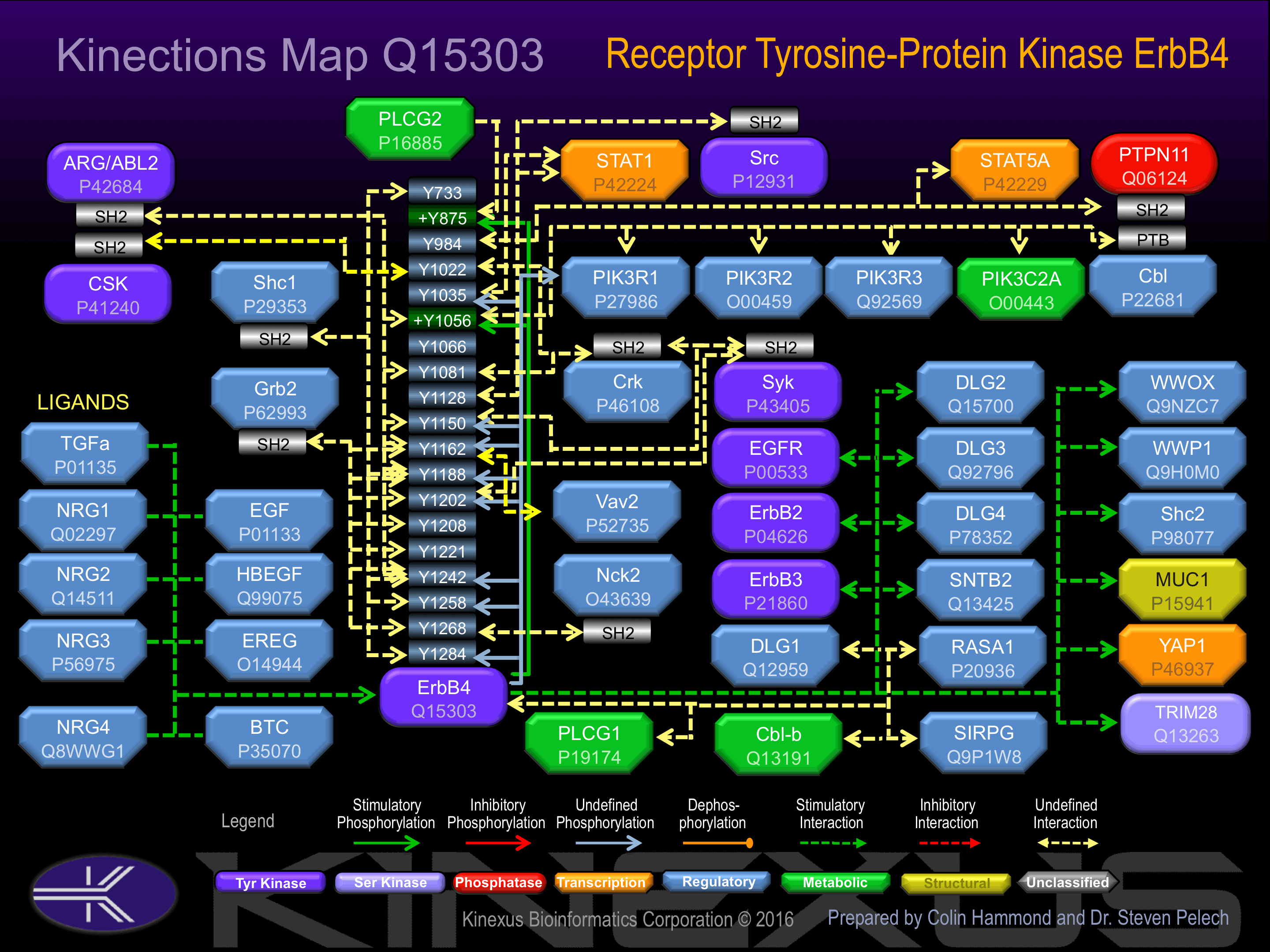
Homodimer or heterodimer with each of the other ERBB receptors Potential. Interacts with PDZ domains of DLG2, DLG3, DLG4 and the syntrophin SNTB2. Interacts with CBFA2T3 and MUC1. Interacts (via PPxy motifs) with WWOX. Isoform JM-A CYT-2 interacts (via PPxY motif 3) with isoform 1 and isoform 2 of YAP1 (via WW domain 1). Isoform JM-A CYT-1 and isoform JM-B CYT-1 interact with WWP1.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
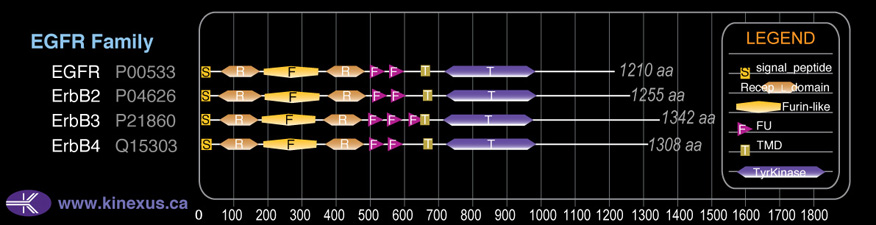
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 25 | signal_peptide |
| 55 | 167 | Recep_L_domain |
| 183 | 335 | Furin-like |
| 358 | 478 | Recep_L_domain |
| 493 | 544 | FU |
| 549 | 599 | FU |
| 653 | 675 | TMD |
| 718 | 974 | TyrKc |
| 718 | 976 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K873, K881.
N-GlcNAcylated:
N138, N174, N181, N253, N358, N410, N473, N495, N548, N576, N620.
Serine phosphorylated:
S726.
Threonine phosphorylated:
T731.
Tyrosine phosphorylated:
Y733, Y875, Y984, Y1022, Y1035, Y1056+, Y1066, Y1081, Y1128, Y1150, Y1162, Y1188, Y1202, Y1208, Y1221, Y1242, Y1258, Y1262, Y1266, Y1268, Y1284, Y1301.
Ubiquitinated:
K722, K745, K858, K1265.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1079
44
1423
 2
2
26
18
30
 5
5
52
11
99
 28
28
300
151
442
 77
77
829
45
675
 5
5
52
98
198
 14
14
149
55
345
 36
36
393
52
722
 24
24
259
17
213
 10
10
112
134
109
 2
2
24
34
30
 48
48
517
213
578
 2
2
20
40
19
 3
3
30
15
34
 3
3
35
31
49
 3
3
27
26
63
 2
2
26
220
130
 8
8
84
29
108
 6
6
65
135
64
 26
26
279
165
242
 8
8
81
32
108
 2
2
21
35
26
 2
2
25
25
43
 5
5
59
29
70
 2
2
22
33
37
 80
80
868
97
1711
 3
3
27
38
28
 5
5
51
28
63
 3
3
30
26
43
 22
22
235
56
154
 22
22
240
24
231
 79
79
849
46
1656
 12
12
131
140
321
 65
65
696
109
634
 9
9
99
61
88
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.9
99.9
99.9
100 48.2
48.2
63.4
- -
-
-
97 -
-
-
- 93.5
93.5
94.8
98 -
-
-
- 96.1
96.1
97.7
97 96.5
96.5
98.4
97 -
-
-
- 88
88
90.6
- 27.5
27.5
36.3
93 -
-
-
84 66
66
76
75 -
-
-
- 35.6
35.6
50.9
- -
-
-
- 20.7
20.7
37.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | ERBB2 - P04626 |
| 2 | ERBB3 - P21860 |
| 3 | DLG3 - Q92796 |
| 4 | NRG2 - O14511 |
| 5 | DLG4 - P78352 |
| 6 | ADAM17 - P78536 |
| 7 | YAP1 - P46937 |
| 8 | TGFA - P01135 |
| 9 | HBEGF - Q99075 |
| 10 | BTC - P35070 |
| 11 | NRG4 - Q8WWG1 |
| 12 | EREG - O14944 |
| 13 | EGFR - P00533 |
| 14 | CD44 - P16070 |
| 15 | CRK - P46108 |
Regulation
Activation:
Phosphorylation at Tyr-1056 induces interaction with PIK3C2A. Phosphorylation at Tyr-1188 induces interaction with Shc1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
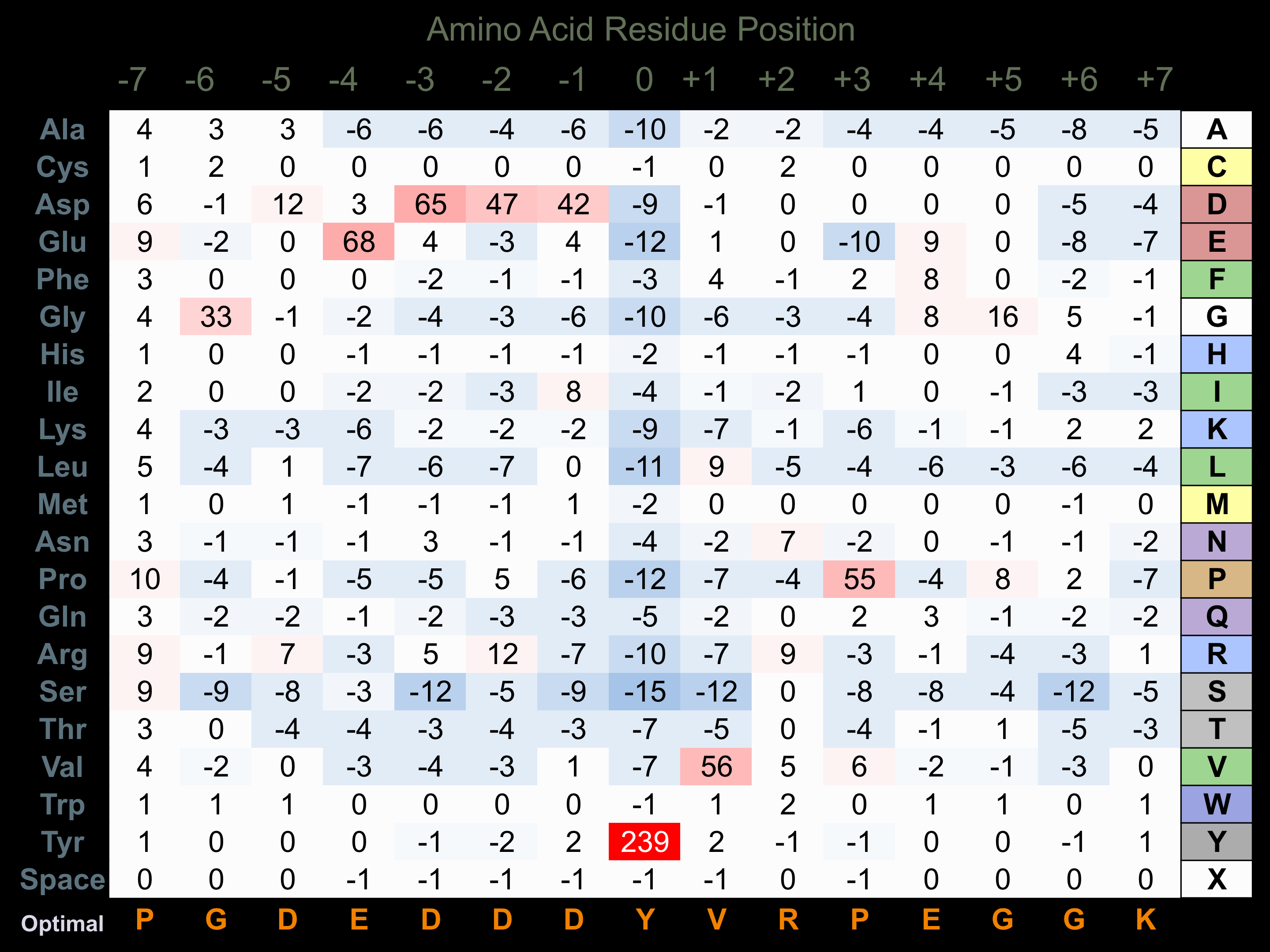
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological, and cardiovascular disorders
Specific Diseases (Non-cancerous):
Amyotrophic lateral sclerosis (ALS); Left Ventricular Outflow tract Obstruction; Early Myoclonic Encephalopathy
Comments:
R927Q and R1275W mutations are associated with amyotrophic lateral sclerosis 19 (ALS19), which is a neurodegenerative disorder affecting upper and lower motor neurons and results in fetal paralysis. Linkage disequilibrium block containing ERBB4 gene was found to be significantly associated with schizophernia.
Specific Cancer Types:
Medulloblastomas; Breast cancer; Colorectal cancer (CRC)
Comments:
ERBB4 is a known oncoprotein (OP) and tumour suppressor protein (TSP). Mutations in ErbB4 are associated with many types of cancer. Multiple missense mutations in ERBB4 have been found in melanoma patients with increased ErbB4 phosphotransferase activity and transformation ability. Melanoma cells with mutant ERBB had reduced cell growth after shRNA knockdown of ERBB4 or treatment with ERBB inhibitor.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -52, p<0.001); Breast epithelial carcinomas (%CFC= -60, p<0.049); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= +104, p<0.089); Cervical cancer stage 2A (%CFC= +212, p<0.01); Cervical cancer stage 2B (%CFC= +307, p<0.0009); Clear cell renal cell carcinomas (cRCC) (%CFC= -49, p<0.024); Malignant pleural mesotheliomas (MPM) tumours (%CFC= -45, p<0.023); Papillary thyroid carcinomas (PTC) (%CFC= -53, p<0.016); Skin melanomas - malignant (%CFC= -54, p<0.02); and Skin squamous cell carcinomas (%CFC= -48, p<0.067). The COSMIC website notes an up-regulated expression score for ERBB4 in diverse human cancers of 393, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.18 % in 25784 diverse cancer specimens. This rate is 2.45-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.08 % in 856 skin cancers tested; 0.87 % in 1128 large intestine cancers tested; 0.6 % in 590 stomach cancers tested; 0.39 % in 1999 lung cancers tested; 0.29 % in 605 oesophagus cancers tested; 0.27 % in 602 endometrium cancers tested; 0.16 % in 984 upper aerodigestive tract cancers tested; 0.16 % in 1270 liver cancers tested; 0.15 % in 500 urinary tract cancers tested; 0.11 % in 1498 breast cancers tested; 0.11 % in 1226 kidney cancers tested; 0.08 % in 1196 pancreas cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: T19A (11).
Comments:
Only 5 deletions, 6 insertions and no complex mutations are noted on the COSMIC website.